Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
Combine predictors using stacking#
Stacking is an ensemble method. In this strategy, the
out-of-fold predictions from several base estimators are used to train a
meta-model that combines their outputs at inference time. Unlike
VotingRegressor, which averages predictions with
fixed (optionally user-specified) weights,
StackingRegressor learns the combination through its
final_estimator.
In this example, we illustrate the use case in which different regressors are stacked together and a final regularized linear regressor is used to output the prediction. We compare the performance of each individual regressor with the stacking strategy. Here, stacking slightly improves the overall performance.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
Generate data#
We use synthetic data generated from a sinusoid plus a linear trend with heteroscedastic Gaussian noise. A sudden drop is introduced, as it cannot be described by a linear model, but a tree-based model can naturally deal with it.
import numpy as np
import pandas as pd
rng = np.random.RandomState(42)
X = rng.uniform(-3, 3, size=500)
trend = 2.4 * X
seasonal = 3.1 * np.sin(3.2 * X)
drop = 10.0 * (X > 2).astype(float)
sigma = 0.75 + 0.75 * X**2
y = trend + seasonal - drop + rng.normal(loc=0.0, scale=np.sqrt(sigma))
df = pd.DataFrame({"X": X, "y": y})
_ = df.plot.scatter(x="X", y="y")
Stack of predictors on a single data set#
It is sometimes not evident which model is more suited for a given task, as
different model families can achieve similar performance while exhibiting
different strengths and weaknesses. Stacking combines their outputs to exploit
these complementary behaviors and can correct systematic errors that no single
model can fix on its own. With appropriate regularization in the
final_estimator, the StackingRegressor often
matches the strongest base model, and can outperform it when base learners’
errors are only partially correlated, allowing the combination to reduce
individual bias/variance.
Here, we combine 3 learners (linear and non-linear) and use the default
RidgeCV regressor to combine their outputs
together.
Note
Although some base learners include preprocessing (such as the
StandardScaler), the final_estimator does
not need additional preprocessing when using the default
passthrough=False, as it receives only the base learners’ predictions. If
passthrough=True, final_estimator should be a pipeline with proper
preprocessing.
from sklearn.ensemble import HistGradientBoostingRegressor, StackingRegressor
from sklearn.linear_model import RidgeCV
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import PolynomialFeatures, SplineTransformer, StandardScaler
linear_ridge = make_pipeline(StandardScaler(), RidgeCV())
spline_ridge = make_pipeline(
SplineTransformer(n_knots=6, degree=3),
PolynomialFeatures(interaction_only=True),
RidgeCV(),
)
hgbt = HistGradientBoostingRegressor(random_state=0)
estimators = [
("Linear Ridge", linear_ridge),
("Spline Ridge", spline_ridge),
("HGBT", hgbt),
]
stacking_regressor = StackingRegressor(estimators=estimators, final_estimator=RidgeCV())
stacking_regressor
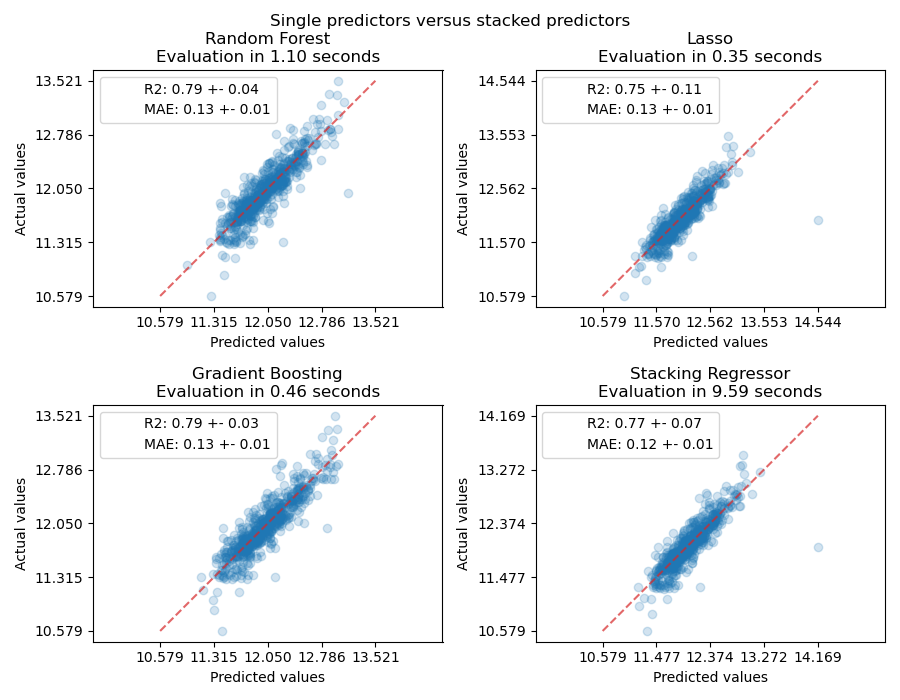
Measure and plot the results#
We can directly plot the predictions. Indeed, the sudden drop is correctly
described by the HistGradientBoostingRegressor
model (HGBT), but the spline model is smoother and less overfitting. The stacked
regressor then turns to be a smoother version of the HGBT.
import matplotlib.pyplot as plt
X = X.reshape(-1, 1)
linear_ridge.fit(X, y)
spline_ridge.fit(X, y)
hgbt.fit(X, y)
stacking_regressor.fit(X, y)
x_plot = np.linspace(X.min() - 0.1, X.max() + 0.1, 500).reshape(-1, 1)
preds = {
"Linear Ridge": linear_ridge.predict(x_plot),
"Spline Ridge": spline_ridge.predict(x_plot),
"HGBT": hgbt.predict(x_plot),
"Stacking (Ridge final estimator)": stacking_regressor.predict(x_plot),
}
fig, axes = plt.subplots(2, 2, figsize=(10, 8), sharex=True, sharey=True)
axes = axes.ravel()
for ax, (name, y_pred) in zip(axes, preds.items()):
ax.scatter(
X[:, 0],
y,
s=6,
alpha=0.35,
linewidths=0,
label="observed (sample)",
)
ax.plot(x_plot.ravel(), y_pred, linewidth=2, alpha=0.9, label=name)
ax.set_title(name)
ax.set_xlabel("x")
ax.set_ylabel("y")
ax.legend(loc="lower right")
plt.suptitle("Base Models Predictions versus Stacked Predictions", y=1)
plt.tight_layout()
plt.show()
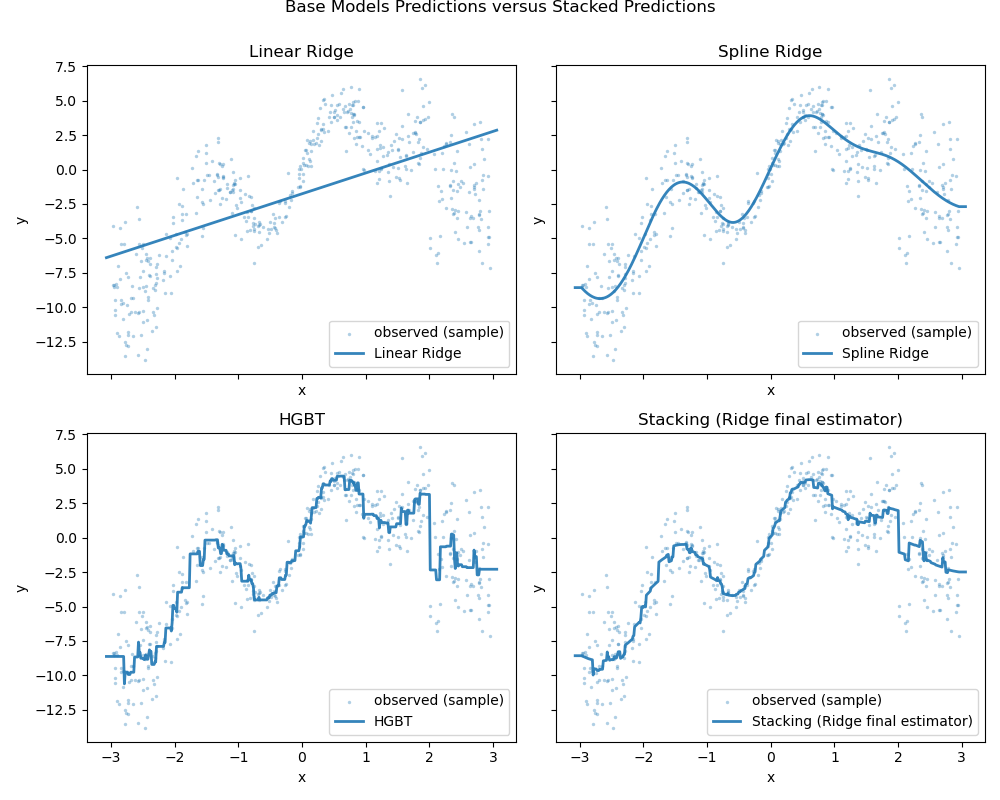
We can plot the prediction errors as well and evaluate the performance of the individual predictors and the stack of the regressors.
import time
from sklearn.metrics import PredictionErrorDisplay
from sklearn.model_selection import cross_val_predict, cross_validate
fig, axs = plt.subplots(2, 2, figsize=(9, 7))
axs = np.ravel(axs)
for ax, (name, est) in zip(
axs, estimators + [("Stacking Regressor", stacking_regressor)]
):
scorers = {r"$R^2$": "r2", "MAE": "neg_mean_absolute_error"}
start_time = time.time()
scores = cross_validate(est, X, y, scoring=list(scorers.values()), n_jobs=-1)
elapsed_time = time.time() - start_time
y_pred = cross_val_predict(est, X, y, n_jobs=-1)
scores = {
key: (
f"{np.abs(np.mean(scores[f'test_{value}'])):.2f}"
r" $\pm$ "
f"{np.std(scores[f'test_{value}']):.2f}"
)
for key, value in scorers.items()
}
display = PredictionErrorDisplay.from_predictions(
y_true=y,
y_pred=y_pred,
kind="actual_vs_predicted",
ax=ax,
scatter_kwargs={"alpha": 0.2, "color": "tab:blue"},
line_kwargs={"color": "tab:red"},
)
ax.set_title(f"{name}\nEvaluation in {elapsed_time:.2f} seconds")
for name, score in scores.items():
ax.plot([], [], " ", label=f"{name}: {score}")
ax.legend(loc="upper left")
plt.suptitle("Prediction Errors of Base versus Stacked Predictors", y=1)
plt.tight_layout()
plt.subplots_adjust(top=0.9)
plt.show()
Even if the scores overlap considerably after cross-validation, the predictions from the stacked regressor are slightly better.
Once fitted, we can inspect the coefficients (or meta-weights) of the trained
final_estimator_ (as long as it is a linear model). They reveal how much the
individual estimators contribute to the the stacked regressor:
stacking_regressor.fit(X, y)
stacking_regressor.final_estimator_.coef_
array([-0.00446216, 0.44878552, 0.54762418])
We see that in this case, the HGBT model dominates, with the spline
ridge also contributing meaningfully. The plain linear model does not add
useful signal once those two are included; with
RidgeCV as the final_estimator, it is not
dropped, but receives a small negative weight to correct its residual bias.
If we use LassoCV as the
final_estimator, that small, unhelpful contribution is set exactly to zero,
yielding a simpler blend of the spline ridge and HGBT models.
from sklearn.linear_model import LassoCV
stacking_regressor = StackingRegressor(estimators=estimators, final_estimator=LassoCV())
stacking_regressor.fit(X, y)
stacking_regressor.final_estimator_.coef_
array([0. , 0.41148006, 0.56187293])
How to mimic SuperLearner with scikit-learn#
The SuperLearner [Polley2010] is a stacking strategy implemented as an R
package, but
not available off-the-shelf in Python. It is closely related to the
StackingRegressor, as both train the meta-model on
out-of-fold predictions from the base estimators.
The key difference is that SuperLearner estimates a convex set of
meta-weights (non-negative and summing to 1) and omits an intercept; by
contrast, StackingRegressor uses an unconstrained
meta-learner with an intercept by default (and can optionally include raw
features via passthrough).
Without an intercept, the meta-weights are directly interpretable as fractional contributions to the final prediction.
from sklearn.linear_model import LinearRegression
linear_reg = LinearRegression(fit_intercept=False, positive=True)
super_learner_like = StackingRegressor(
estimators=estimators, final_estimator=linear_reg
)
super_learner_like.fit(X, y)
super_learner_like.final_estimator_.coef_
array([2.41599724e-04, 4.48129539e-01, 5.49327451e-01])
The sum of meta-weights in the stacked regressor is close to 1.0, but not exactly one:
super_learner_like.final_estimator_.coef_.sum()
np.float64(0.9976985896404211)
Beyond interpretability, the normalization to 1.0 constraint in the SuperLearner
presents the following advantages:
Consensus-preserving: if all base models output the same value at a point, the ensemble returns that same value (no artificial amplification or attenuation).
Translation-equivariant: adding a constant to every base prediction shifts the ensemble by the same constant.
Removes one degree of freedom: avoiding redundancy with a constant term and modestly stabilizing weights under collinearity.
The cleanest way to enforce the coefficient normalization with scikit-learn is by defining a custom estimator, but doing so is beyond the scope of this tutorial.
Conclusions#
The stacked regressor combines the strengths of the different regressors. However, notice that training the stacked regressor is much more computationally expensive than selecting the best performing model.
References
Polley, E. C. and van der Laan, M. J., Super Learner In Prediction, 2010.
Total running time of the script: (0 minutes 5.844 seconds)
Related examples