Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
Demo of OPTICS clustering algorithm#
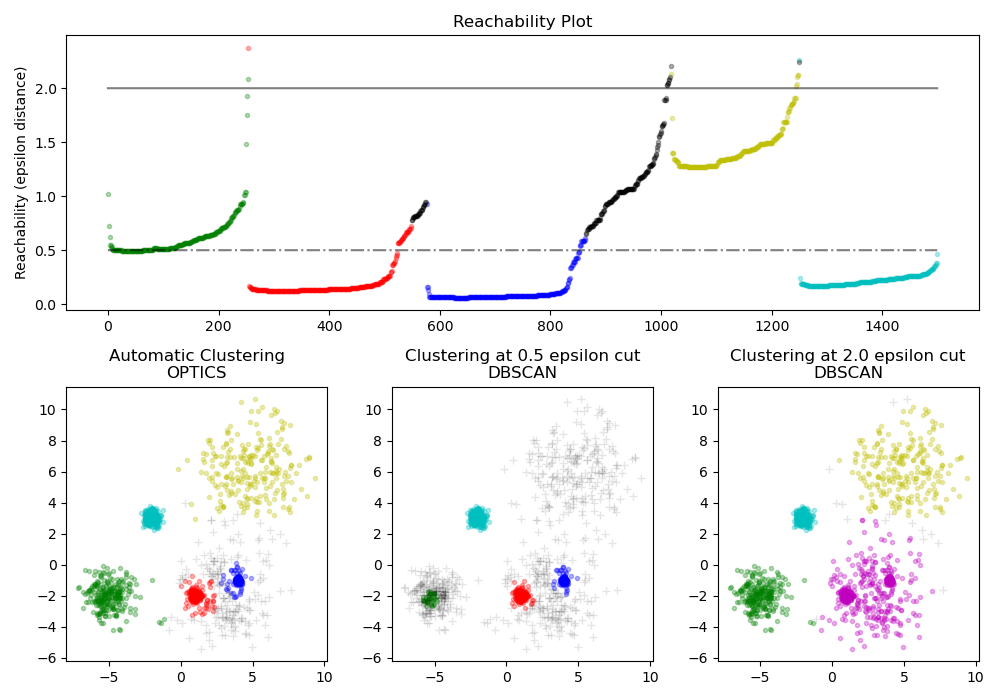
Finds core samples of high density and expands clusters from them. This example uses data that is generated so that the clusters have different densities.
The OPTICS is first used with its Xi cluster detection
method, and then setting specific thresholds on the reachability, which
corresponds to DBSCAN. We can see that the different
clusters of OPTICS’s Xi method can be recovered with different choices of
thresholds in DBSCAN.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import matplotlib.gridspec as gridspec
import matplotlib.pyplot as plt
import numpy as np
from sklearn.cluster import OPTICS, cluster_optics_dbscan
# Generate sample data
np.random.seed(0)
n_points_per_cluster = 250
C1 = [-5, -2] + 0.8 * np.random.randn(n_points_per_cluster, 2)
C2 = [4, -1] + 0.1 * np.random.randn(n_points_per_cluster, 2)
C3 = [1, -2] + 0.2 * np.random.randn(n_points_per_cluster, 2)
C4 = [-2, 3] + 0.3 * np.random.randn(n_points_per_cluster, 2)
C5 = [3, -2] + 1.6 * np.random.randn(n_points_per_cluster, 2)
C6 = [5, 6] + 2 * np.random.randn(n_points_per_cluster, 2)
X = np.vstack((C1, C2, C3, C4, C5, C6))
clust = OPTICS(min_samples=50, xi=0.05, min_cluster_size=0.05)
# Run the fit
clust.fit(X)
labels_050 = cluster_optics_dbscan(
reachability=clust.reachability_,
core_distances=clust.core_distances_,
ordering=clust.ordering_,
eps=0.5,
)
labels_200 = cluster_optics_dbscan(
reachability=clust.reachability_,
core_distances=clust.core_distances_,
ordering=clust.ordering_,
eps=2,
)
space = np.arange(len(X))
reachability = clust.reachability_[clust.ordering_]
labels = clust.labels_[clust.ordering_]
plt.figure(figsize=(10, 7))
G = gridspec.GridSpec(2, 3)
ax1 = plt.subplot(G[0, :])
ax2 = plt.subplot(G[1, 0])
ax3 = plt.subplot(G[1, 1])
ax4 = plt.subplot(G[1, 2])
# Reachability plot
colors = ["g.", "r.", "b.", "y.", "c."]
for klass, color in enumerate(colors):
Xk = space[labels == klass]
Rk = reachability[labels == klass]
ax1.plot(Xk, Rk, color, alpha=0.3)
ax1.plot(space[labels == -1], reachability[labels == -1], "k.", alpha=0.3)
ax1.plot(space, np.full_like(space, 2.0, dtype=float), "k-", alpha=0.5)
ax1.plot(space, np.full_like(space, 0.5, dtype=float), "k-.", alpha=0.5)
ax1.set_ylabel("Reachability (epsilon distance)")
ax1.set_title("Reachability Plot")
# OPTICS
colors = ["g.", "r.", "b.", "y.", "c."]
for klass, color in enumerate(colors):
Xk = X[clust.labels_ == klass]
ax2.plot(Xk[:, 0], Xk[:, 1], color, alpha=0.3)
ax2.plot(X[clust.labels_ == -1, 0], X[clust.labels_ == -1, 1], "k+", alpha=0.1)
ax2.set_title("Automatic Clustering\nOPTICS")
# DBSCAN at 0.5
colors = ["g.", "r.", "b.", "c."]
for klass, color in enumerate(colors):
Xk = X[labels_050 == klass]
ax3.plot(Xk[:, 0], Xk[:, 1], color, alpha=0.3)
ax3.plot(X[labels_050 == -1, 0], X[labels_050 == -1, 1], "k+", alpha=0.1)
ax3.set_title("Clustering at 0.5 epsilon cut\nDBSCAN")
# DBSCAN at 2.
colors = ["g.", "m.", "y.", "c."]
for klass, color in enumerate(colors):
Xk = X[labels_200 == klass]
ax4.plot(Xk[:, 0], Xk[:, 1], color, alpha=0.3)
ax4.plot(X[labels_200 == -1, 0], X[labels_200 == -1, 1], "k+", alpha=0.1)
ax4.set_title("Clustering at 2.0 epsilon cut\nDBSCAN")
plt.tight_layout()
plt.show()
Total running time of the script: (0 minutes 1.465 seconds)
Related examples
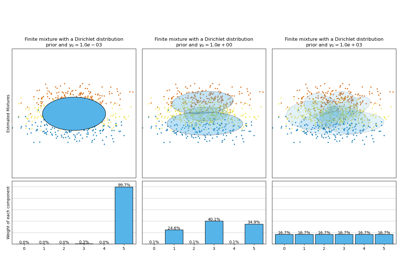
Concentration Prior Type Analysis of Variation Bayesian Gaussian Mixture
Concentration Prior Type Analysis of Variation Bayesian Gaussian Mixture