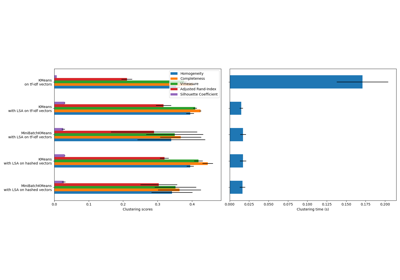
Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
Wikipedia principal eigenvector#
A classical way to assert the relative importance of vertices in a graph is to compute the principal eigenvector of the adjacency matrix so as to assign to each vertex the values of the components of the first eigenvector as a centrality score: https://en.wikipedia.org/wiki/Eigenvector_centrality. On the graph of webpages and links those values are called the PageRank scores by Google.
The goal of this example is to analyze the graph of links inside wikipedia articles to rank articles by relative importance according to this eigenvector centrality.
The traditional way to compute the principal eigenvector is to use the power iteration method. Here the computation is achieved thanks to Martinsson’s Randomized SVD algorithm implemented in scikit-learn.
The graph data is fetched from the DBpedia dumps. DBpedia is an extraction of the latent structured data of the Wikipedia content.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import os
from bz2 import BZ2File
from datetime import datetime
from pprint import pprint
from time import time
from urllib.request import urlopen
import numpy as np
from scipy import sparse
from sklearn.decomposition import randomized_svd
Download data, if not already on disk#
redirects_url = "http://downloads.dbpedia.org/3.5.1/en/redirects_en.nt.bz2"
redirects_filename = redirects_url.rsplit("/", 1)[1]
page_links_url = "http://downloads.dbpedia.org/3.5.1/en/page_links_en.nt.bz2"
page_links_filename = page_links_url.rsplit("/", 1)[1]
resources = [
(redirects_url, redirects_filename),
(page_links_url, page_links_filename),
]
for url, filename in resources:
if not os.path.exists(filename):
print("Downloading data from '%s', please wait..." % url)
opener = urlopen(url)
with open(filename, "wb") as f:
f.write(opener.read())
print()
Loading the redirect files#
def index(redirects, index_map, k):
"""Find the index of an article name after redirect resolution"""
k = redirects.get(k, k)
return index_map.setdefault(k, len(index_map))
DBPEDIA_RESOURCE_PREFIX_LEN = len("http://dbpedia.org/resource/")
SHORTNAME_SLICE = slice(DBPEDIA_RESOURCE_PREFIX_LEN + 1, -1)
def short_name(nt_uri):
"""Remove the < and > URI markers and the common URI prefix"""
return nt_uri[SHORTNAME_SLICE]
def get_redirects(redirects_filename):
"""Parse the redirections and build a transitively closed map out of it"""
redirects = {}
print("Parsing the NT redirect file")
for l, line in enumerate(BZ2File(redirects_filename)):
split = line.split()
if len(split) != 4:
print("ignoring malformed line: " + line)
continue
redirects[short_name(split[0])] = short_name(split[2])
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
# compute the transitive closure
print("Computing the transitive closure of the redirect relation")
for l, source in enumerate(redirects.keys()):
transitive_target = None
target = redirects[source]
seen = {source}
while True:
transitive_target = target
target = redirects.get(target)
if target is None or target in seen:
break
seen.add(target)
redirects[source] = transitive_target
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
return redirects
Computing the Adjacency matrix#
def get_adjacency_matrix(redirects_filename, page_links_filename, limit=None):
"""Extract the adjacency graph as a scipy sparse matrix
Redirects are resolved first.
Returns X, the scipy sparse adjacency matrix, redirects as python
dict from article names to article names and index_map a python dict
from article names to python int (article indexes).
"""
print("Computing the redirect map")
redirects = get_redirects(redirects_filename)
print("Computing the integer index map")
index_map = dict()
links = list()
for l, line in enumerate(BZ2File(page_links_filename)):
split = line.split()
if len(split) != 4:
print("ignoring malformed line: " + line)
continue
i = index(redirects, index_map, short_name(split[0]))
j = index(redirects, index_map, short_name(split[2]))
links.append((i, j))
if l % 1000000 == 0:
print("[%s] line: %08d" % (datetime.now().isoformat(), l))
if limit is not None and l >= limit - 1:
break
print("Computing the adjacency matrix")
X = sparse.lil_matrix((len(index_map), len(index_map)), dtype=np.float32)
for i, j in links:
X[i, j] = 1.0
del links
print("Converting to CSR representation")
X = X.tocsr()
print("CSR conversion done")
return X, redirects, index_map
# stop after 5M links to make it possible to work in RAM
X, redirects, index_map = get_adjacency_matrix(
redirects_filename, page_links_filename, limit=5000000
)
names = {i: name for name, i in index_map.items()}
Computing Principal Singular Vector using Randomized SVD#
print("Computing the principal singular vectors using randomized_svd")
t0 = time()
U, s, V = randomized_svd(X, 5, n_iter=3)
print("done in %0.3fs" % (time() - t0))
# print the names of the wikipedia related strongest components of the
# principal singular vector which should be similar to the highest eigenvector
print("Top wikipedia pages according to principal singular vectors")
pprint([names[i] for i in np.abs(U.T[0]).argsort()[-10:]])
pprint([names[i] for i in np.abs(V[0]).argsort()[-10:]])
Computing Centrality scores#
def centrality_scores(X, alpha=0.85, max_iter=100, tol=1e-10):
"""Power iteration computation of the principal eigenvector
This method is also known as Google PageRank and the implementation
is based on the one from the NetworkX project (BSD licensed too)
with copyrights by:
Aric Hagberg <hagberg@lanl.gov>
Dan Schult <dschult@colgate.edu>
Pieter Swart <swart@lanl.gov>
"""
n = X.shape[0]
X = X.copy()
incoming_counts = np.asarray(X.sum(axis=1)).ravel()
print("Normalizing the graph")
for i in incoming_counts.nonzero()[0]:
X.data[X.indptr[i] : X.indptr[i + 1]] *= 1.0 / incoming_counts[i]
dangle = np.asarray(np.where(np.isclose(X.sum(axis=1), 0), 1.0 / n, 0)).ravel()
scores = np.full(n, 1.0 / n, dtype=np.float32) # initial guess
for i in range(max_iter):
print("power iteration #%d" % i)
prev_scores = scores
scores = (
alpha * (scores * X + np.dot(dangle, prev_scores))
+ (1 - alpha) * prev_scores.sum() / n
)
# check convergence: normalized l_inf norm
scores_max = np.abs(scores).max()
if scores_max == 0.0:
scores_max = 1.0
err = np.abs(scores - prev_scores).max() / scores_max
print("error: %0.6f" % err)
if err < n * tol:
return scores
return scores
print("Computing principal eigenvector score using a power iteration method")
t0 = time()
scores = centrality_scores(X, max_iter=100)
print("done in %0.3fs" % (time() - t0))
pprint([names[i] for i in np.abs(scores).argsort()[-10:]])
Related examples
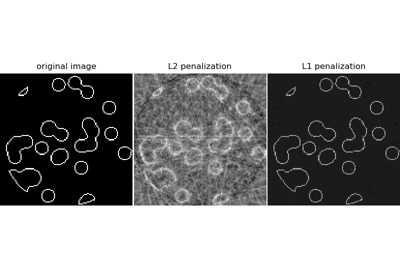
Compressive sensing: tomography reconstruction with L1 prior (Lasso)
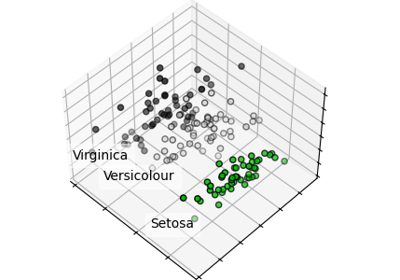
Principal Component Analysis (PCA) on Iris Dataset