Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
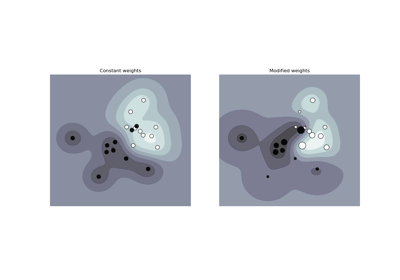
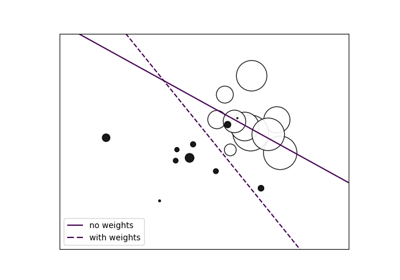
Fitting an Elastic Net with a precomputed Gram Matrix and Weighted Samples#
The following example shows how to precompute the gram matrix
while using weighted samples with an ElasticNet.
If weighted samples are used, the design matrix must be centered and then rescaled by the square root of the weight vector before the gram matrix is computed.
Note
sample_weightvector is also rescaled to sum ton_samples, see thedocumentation for the
sample_weightparameter tofit.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
Let’s start by loading the dataset and creating some sample weights.
import numpy as np
from sklearn.datasets import make_regression
rng = np.random.RandomState(0)
n_samples = int(1e5)
X, y = make_regression(n_samples=n_samples, noise=0.5, random_state=rng)
sample_weight = rng.lognormal(size=n_samples)
# normalize the sample weights
normalized_weights = sample_weight * (n_samples / (sample_weight.sum()))
To fit the elastic net using the precompute option together with the sample
weights, we must first center the design matrix, and rescale it by the
normalized weights prior to computing the gram matrix.
X_offset = np.average(X, axis=0, weights=normalized_weights)
X_centered = X - np.average(X, axis=0, weights=normalized_weights)
X_scaled = X_centered * np.sqrt(normalized_weights)[:, np.newaxis]
gram = np.dot(X_scaled.T, X_scaled)
We can now proceed with fitting. We must passed the centered design matrix to
fit otherwise the elastic net estimator will detect that it is uncentered
and discard the gram matrix we passed. However, if we pass the scaled design
matrix, the preprocessing code will incorrectly rescale it a second time.
from sklearn.linear_model import ElasticNet
lm = ElasticNet(alpha=0.01, precompute=gram)
lm.fit(X_centered, y, sample_weight=normalized_weights)
Total running time of the script: (0 minutes 2.329 seconds)
Related examples
Developing Estimators Compliant with Metadata Routing