Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
Gradient Boosting regression#
This example demonstrates Gradient Boosting to produce a predictive
model from an ensemble of weak predictive models. Gradient boosting can be used
for regression and classification problems. Here, we will train a model to
tackle a diabetes regression task. We will obtain the results from
GradientBoostingRegressor with least squares loss
and 500 regression trees of depth 4.
Note: For larger datasets (n_samples >= 10000), please refer to
HistGradientBoostingRegressor. See
Features in Histogram Gradient Boosting Trees for an example
showcasing some other advantages of
HistGradientBoostingRegressor.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets, ensemble
from sklearn.inspection import permutation_importance
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.utils.fixes import parse_version
Load the data#
First we need to load the data.
diabetes = datasets.load_diabetes()
X, y = diabetes.data, diabetes.target
Data preprocessing#
Next, we will split our dataset to use 90% for training and leave the rest for testing. We will also set the regression model parameters. You can play with these parameters to see how the results change.
n_estimators : the number of boosting stages that will be performed.
Later, we will plot deviance against boosting iterations.
max_depth : limits the number of nodes in the tree.
The best value depends on the interaction of the input variables.
min_samples_split : the minimum number of samples required to split an
internal node.
learning_rate : how much the contribution of each tree will shrink.
loss : loss function to optimize. The least squares function is used in
this case however, there are many other options (see
GradientBoostingRegressor ).
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=13
)
params = {
"n_estimators": 500,
"max_depth": 4,
"min_samples_split": 5,
"learning_rate": 0.01,
"loss": "squared_error",
}
Fit regression model#
Now we will initiate the gradient boosting regressors and fit it with our training data. Let’s also look and the mean squared error on the test data.
reg = ensemble.GradientBoostingRegressor(**params)
reg.fit(X_train, y_train)
mse = mean_squared_error(y_test, reg.predict(X_test))
print("The mean squared error (MSE) on test set: {:.4f}".format(mse))
The mean squared error (MSE) on test set: 3010.2061
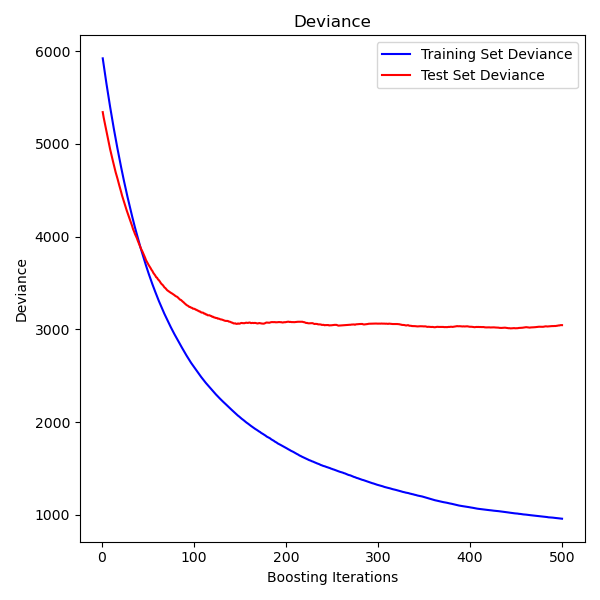
Plot training deviance#
Finally, we will visualize the results. To do that we will first compute the test set deviance and then plot it against boosting iterations.
test_score = np.zeros((params["n_estimators"],), dtype=np.float64)
for i, y_pred in enumerate(reg.staged_predict(X_test)):
test_score[i] = mean_squared_error(y_test, y_pred)
fig = plt.figure(figsize=(6, 6))
plt.subplot(1, 1, 1)
plt.title("Deviance")
plt.plot(
np.arange(params["n_estimators"]) + 1,
reg.train_score_,
"b-",
label="Training Set Deviance",
)
plt.plot(
np.arange(params["n_estimators"]) + 1, test_score, "r-", label="Test Set Deviance"
)
plt.legend(loc="upper right")
plt.xlabel("Boosting Iterations")
plt.ylabel("Deviance")
fig.tight_layout()
plt.show()
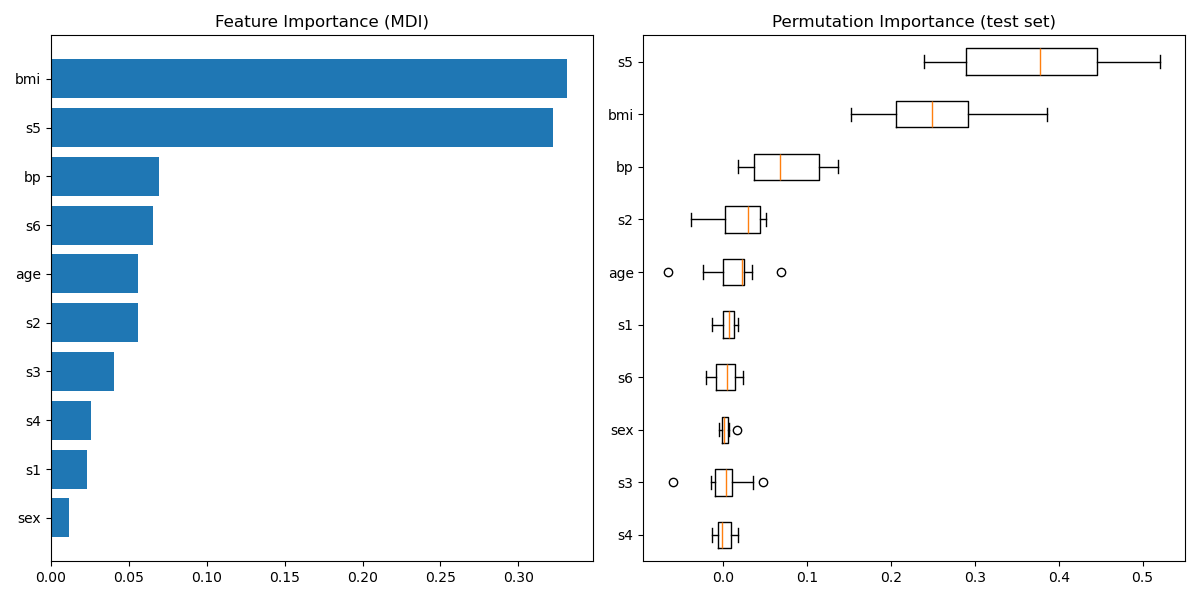
Plot feature importance#
Warning
Careful, impurity-based feature importances can be misleading for
high cardinality features (many unique values). As an alternative,
the permutation importances of reg can be computed on a
held out test set. See Permutation feature importance for more details.
For this example, the impurity-based and permutation methods identify the same 2 strongly predictive features but not in the same order. The third most predictive feature, “bp”, is also the same for the 2 methods. The remaining features are less predictive and the error bars of the permutation plot show that they overlap with 0.
feature_importance = reg.feature_importances_
sorted_idx = np.argsort(feature_importance)
pos = np.arange(sorted_idx.shape[0]) + 0.5
fig = plt.figure(figsize=(12, 6))
plt.subplot(1, 2, 1)
plt.barh(pos, feature_importance[sorted_idx], align="center")
plt.yticks(pos, np.array(diabetes.feature_names)[sorted_idx])
plt.title("Feature Importance (MDI)")
result = permutation_importance(
reg, X_test, y_test, n_repeats=10, random_state=42, n_jobs=2
)
sorted_idx = result.importances_mean.argsort()
plt.subplot(1, 2, 2)
# `labels` argument in boxplot is deprecated in matplotlib 3.9 and has been
# renamed to `tick_labels`. The following code handles this, but as a
# scikit-learn user you probably can write simpler code by using `labels=...`
# (matplotlib < 3.9) or `tick_labels=...` (matplotlib >= 3.9).
tick_labels_parameter_name = (
"tick_labels"
if parse_version(matplotlib.__version__) >= parse_version("3.9")
else "labels"
)
tick_labels_dict = {
tick_labels_parameter_name: np.array(diabetes.feature_names)[sorted_idx]
}
plt.boxplot(result.importances[sorted_idx].T, vert=False, **tick_labels_dict)
plt.title("Permutation Importance (test set)")
fig.tight_layout()
plt.show()
Total running time of the script: (0 minutes 1.247 seconds)
Related examples
Permutation Importance vs Random Forest Feature Importance (MDI)
Permutation Importance with Multicollinear or Correlated Features