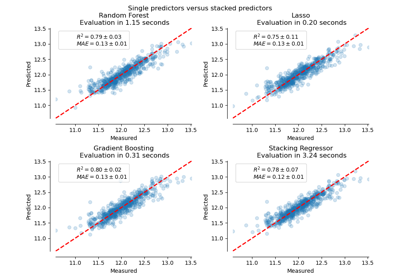
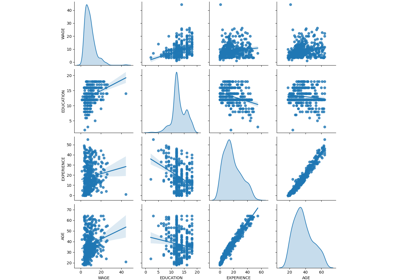
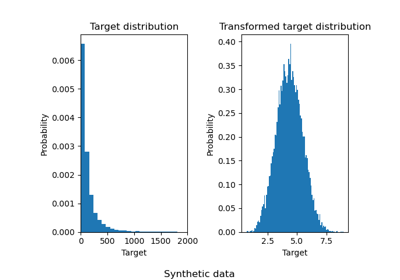
3.2.4.1.9. sklearn.linear_model.RidgeCV¶
-
class
sklearn.linear_model.RidgeCV(alphas=(0.1, 1.0, 10.0), *, fit_intercept=True, normalize=False, scoring=None, cv=None, gcv_mode=None, store_cv_values=False)[source]¶ Ridge regression with built-in cross-validation.
See glossary entry for cross-validation estimator.
By default, it performs Generalized Cross-Validation, which is a form of efficient Leave-One-Out cross-validation.
Read more in the User Guide.
- Parameters
- alphasndarray of shape (n_alphas,), default=(0.1, 1.0, 10.0)
Array of alpha values to try. Regularization strength; must be a positive float. Regularization improves the conditioning of the problem and reduces the variance of the estimates. Larger values specify stronger regularization. Alpha corresponds to
1 / (2C)in other linear models such asLogisticRegressionorsklearn.svm.LinearSVC. If using generalized cross-validation, alphas must be positive.- fit_interceptbool, default=True
Whether to calculate the intercept for this model. If set to false, no intercept will be used in calculations (i.e. data is expected to be centered).
- normalizebool, default=False
This parameter is ignored when
fit_interceptis set to False. If True, the regressors X will be normalized before regression by subtracting the mean and dividing by the l2-norm. If you wish to standardize, please usesklearn.preprocessing.StandardScalerbefore callingfiton an estimator withnormalize=False.- scoringstring, callable, default=None
A string (see model evaluation documentation) or a scorer callable object / function with signature
scorer(estimator, X, y). If None, the negative mean squared error if cv is ‘auto’ or None (i.e. when using generalized cross-validation), and r2 score otherwise.- cvint, cross-validation generator or an iterable, default=None
Determines the cross-validation splitting strategy. Possible inputs for cv are:
None, to use the efficient Leave-One-Out cross-validation (also known as Generalized Cross-Validation).
integer, to specify the number of folds.
An iterable yielding (train, test) splits as arrays of indices.
For integer/None inputs, if
yis binary or multiclass,sklearn.model_selection.StratifiedKFoldis used, else,sklearn.model_selection.KFoldis used.Refer User Guide for the various cross-validation strategies that can be used here.
- gcv_mode{‘auto’, ‘svd’, eigen’}, default=’auto’
Flag indicating which strategy to use when performing Generalized Cross-Validation. Options are:
'auto' : use 'svd' if n_samples > n_features, otherwise use 'eigen' 'svd' : force use of singular value decomposition of X when X is dense, eigenvalue decomposition of X^T.X when X is sparse. 'eigen' : force computation via eigendecomposition of X.X^T
The ‘auto’ mode is the default and is intended to pick the cheaper option of the two depending on the shape of the training data.
- store_cv_valuesbool, default=False
Flag indicating if the cross-validation values corresponding to each alpha should be stored in the
cv_values_attribute (see below). This flag is only compatible withcv=None(i.e. using Generalized Cross-Validation).
- Attributes
- cv_values_ndarray of shape (n_samples, n_alphas) or shape (n_samples, n_targets, n_alphas), optional
Cross-validation values for each alpha (only available if
store_cv_values=Trueandcv=None). Afterfit()has been called, this attribute will contain the mean squared errors (by default) or the values of the{loss,score}_funcfunction (if provided in the constructor).- coef_ndarray of shape (n_features) or (n_targets, n_features)
Weight vector(s).
- intercept_float or ndarray of shape (n_targets,)
Independent term in decision function. Set to 0.0 if
fit_intercept = False.- alpha_float
Estimated regularization parameter.
- best_score_float
Score of base estimator with best alpha.
See also
RidgeRidge regression
RidgeClassifierRidge classifier
RidgeClassifierCVRidge classifier with built-in cross validation
Examples
>>> from sklearn.datasets import load_diabetes >>> from sklearn.linear_model import RidgeCV >>> X, y = load_diabetes(return_X_y=True) >>> clf = RidgeCV(alphas=[1e-3, 1e-2, 1e-1, 1]).fit(X, y) >>> clf.score(X, y) 0.5166...
Methods
fit(X, y[, sample_weight])Fit Ridge regression model with cv.
get_params([deep])Get parameters for this estimator.
predict(X)Predict using the linear model.
score(X, y[, sample_weight])Return the coefficient of determination R^2 of the prediction.
set_params(**params)Set the parameters of this estimator.
-
__init__(alphas=(0.1, 1.0, 10.0), *, fit_intercept=True, normalize=False, scoring=None, cv=None, gcv_mode=None, store_cv_values=False)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(X, y, sample_weight=None)[source]¶ Fit Ridge regression model with cv.
- Parameters
- Xndarray of shape (n_samples, n_features)
Training data. If using GCV, will be cast to float64 if necessary.
- yndarray of shape (n_samples,) or (n_samples, n_targets)
Target values. Will be cast to X’s dtype if necessary.
- sample_weightfloat or ndarray of shape (n_samples,), default=None
Individual weights for each sample. If given a float, every sample will have the same weight.
- Returns
- selfobject
Notes
When sample_weight is provided, the selected hyperparameter may depend on whether we use generalized cross-validation (cv=None or cv=’auto’) or another form of cross-validation, because only generalized cross-validation takes the sample weights into account when computing the validation score.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsmapping of string to any
Parameter names mapped to their values.
-
predict(X)[source]¶ Predict using the linear model.
- Parameters
- Xarray_like or sparse matrix, shape (n_samples, n_features)
Samples.
- Returns
- Carray, shape (n_samples,)
Returns predicted values.
-
score(X, y, sample_weight=None)[source]¶ Return the coefficient of determination R^2 of the prediction.
The coefficient R^2 is defined as (1 - u/v), where u is the residual sum of squares ((y_true - y_pred) ** 2).sum() and v is the total sum of squares ((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a R^2 score of 0.0.
- Parameters
- Xarray-like of shape (n_samples, n_features)
Test samples. For some estimators this may be a precomputed kernel matrix or a list of generic objects instead, shape = (n_samples, n_samples_fitted), where n_samples_fitted is the number of samples used in the fitting for the estimator.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True values for X.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns
- scorefloat
R^2 of self.predict(X) wrt. y.
Notes
The R2 score used when calling
scoreon a regressor usesmultioutput='uniform_average'from version 0.23 to keep consistent with default value ofr2_score. This influences thescoremethod of all the multioutput regressors (except forMultiOutputRegressor).
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfobject
Estimator instance.