Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
Release Highlights for scikit-learn 1.0¶
We are very pleased to announce the release of scikit-learn 1.0! The library has been stable for quite some time, releasing version 1.0 is recognizing that and signalling it to our users. This release does not include any breaking changes apart from the usual two-release deprecation cycle. For the future, we do our best to keep this pattern.
This release includes some new key features as well as many improvements and bug fixes. We detail below a few of the major features of this release. For an exhaustive list of all the changes, please refer to the release notes.
To install the latest version (with pip):
pip install --upgrade scikit-learn
or with conda:
conda install -c conda-forge scikit-learn
Keyword and positional arguments¶
The scikit-learn API exposes many functions and methods which have many input
parameters. For example, before this release, one could instantiate a
HistGradientBoostingRegressor as:
HistGradientBoostingRegressor("squared_error", 0.1, 100, 31, None,
20, 0.0, 255, None, None, False, "auto", "loss", 0.1, 10, 1e-7,
0, None)
Understanding the above code requires the reader to go to the API documentation and to check each and every parameter for its position and its meaning. To improve the readability of code written based on scikit-learn, now users have to provide most parameters with their names, as keyword arguments, instead of positional arguments. For example, the above code would be:
HistGradientBoostingRegressor(
loss="squared_error",
learning_rate=0.1,
max_iter=100,
max_leaf_nodes=31,
max_depth=None,
min_samples_leaf=20,
l2_regularization=0.0,
max_bins=255,
categorical_features=None,
monotonic_cst=None,
warm_start=False,
early_stopping="auto",
scoring="loss",
validation_fraction=0.1,
n_iter_no_change=10,
tol=1e-7,
verbose=0,
random_state=None,
)
which is much more readable. Positional arguments have been deprecated since
version 0.23 and will now raise a TypeError. A limited number of
positional arguments are still allowed in some cases, for example in
PCA, where PCA(10) is still allowed, but PCA(10,
False) is not allowed.
Spline Transformers¶
One way to add nonlinear terms to a dataset’s feature set is to generate
spline basis functions for continuous/numerical features with the new
SplineTransformer. Splines are piecewise polynomials,
parametrized by their polynomial degree and the positions of the knots. The
SplineTransformer implements a B-spline basis.
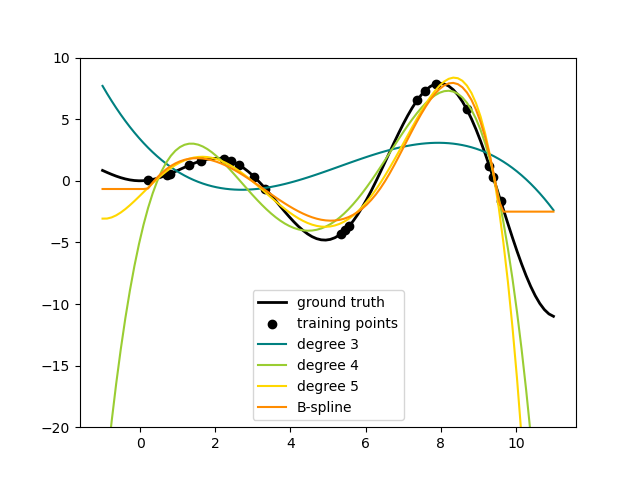
The following code shows splines in action, for more information, please refer to the User Guide.
import numpy as np
from sklearn.preprocessing import SplineTransformer
X = np.arange(5).reshape(5, 1)
spline = SplineTransformer(degree=2, n_knots=3)
spline.fit_transform(X)
array([[0.5 , 0.5 , 0. , 0. ],
[0.125, 0.75 , 0.125, 0. ],
[0. , 0.5 , 0.5 , 0. ],
[0. , 0.125, 0.75 , 0.125],
[0. , 0. , 0.5 , 0.5 ]])
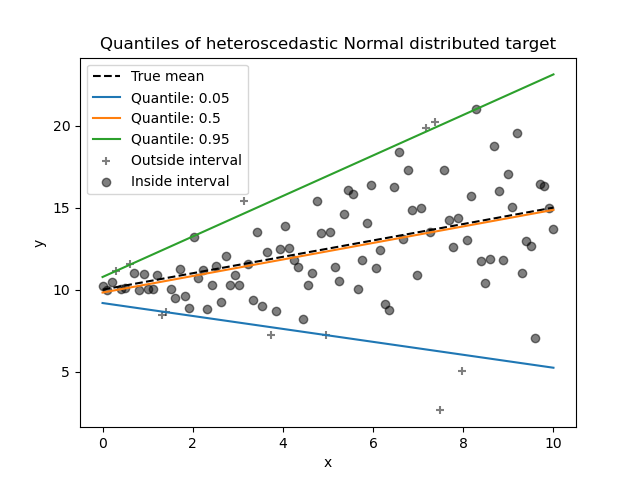
Quantile Regressor¶
Quantile regression estimates the median or other quantiles of \(y\) conditional on \(X\), while ordinary least squares (OLS) estimates the conditional mean.
As a linear model, the new QuantileRegressor gives
linear predictions \(\hat{y}(w, X) = Xw\) for the \(q\)-th quantile,
\(q \in (0, 1)\). The weights or coefficients \(w\) are then found by
the following minimization problem:
This consists of the pinball loss (also known as linear loss),
see also mean_pinball_loss,
and the L1 penalty controlled by parameter alpha, similar to
linear_model.Lasso.
Please check the following example to see how it works, and the User Guide for more details.
Feature Names Support¶
When an estimator is passed a pandas’ dataframe during
fit, the estimator will set a feature_names_in_ attribute
containing the feature names. Note that feature names support is only enabled
when the column names in the dataframe are all strings. feature_names_in_
is used to check that the column names of the dataframe passed in
non-fit, such as predict, are consistent with features in
fit:
from sklearn.preprocessing import StandardScaler
import pandas as pd
X = pd.DataFrame([[1, 2, 3], [4, 5, 6]], columns=["a", "b", "c"])
scalar = StandardScaler().fit(X)
scalar.feature_names_in_
array(['a', 'b', 'c'], dtype=object)
The support of get_feature_names_out is available for transformers
that already had get_feature_names and transformers with a one-to-one
correspondence between input and output such as
StandardScaler. get_feature_names_out support
will be added to all other transformers in future releases. Additionally,
compose.ColumnTransformer.get_feature_names_out is available to
combine feature names of its transformers:
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
import pandas as pd
X = pd.DataFrame({"pet": ["dog", "cat", "fish"], "age": [3, 7, 1]})
preprocessor = ColumnTransformer(
[
("numerical", StandardScaler(), ["age"]),
("categorical", OneHotEncoder(), ["pet"]),
],
verbose_feature_names_out=False,
).fit(X)
preprocessor.get_feature_names_out()
array(['age', 'pet_cat', 'pet_dog', 'pet_fish'], dtype=object)
When this preprocessor is used with a pipeline, the feature names used
by the classifier are obtained by slicing and calling
get_feature_names_out:
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
y = [1, 0, 1]
pipe = make_pipeline(preprocessor, LogisticRegression())
pipe.fit(X, y)
pipe[:-1].get_feature_names_out()
array(['age', 'pet_cat', 'pet_dog', 'pet_fish'], dtype=object)
A more flexible plotting API¶
metrics.ConfusionMatrixDisplay,
metrics.PrecisionRecallDisplay, metrics.DetCurveDisplay,
and inspection.PartialDependenceDisplay now expose two class
methods: from_estimator and from_predictions which allow users to create
a plot given the predictions or an estimator. This means the corresponding
plot_* functions are deprecated. Please check example one and
example two for
how to use the new plotting functionalities.
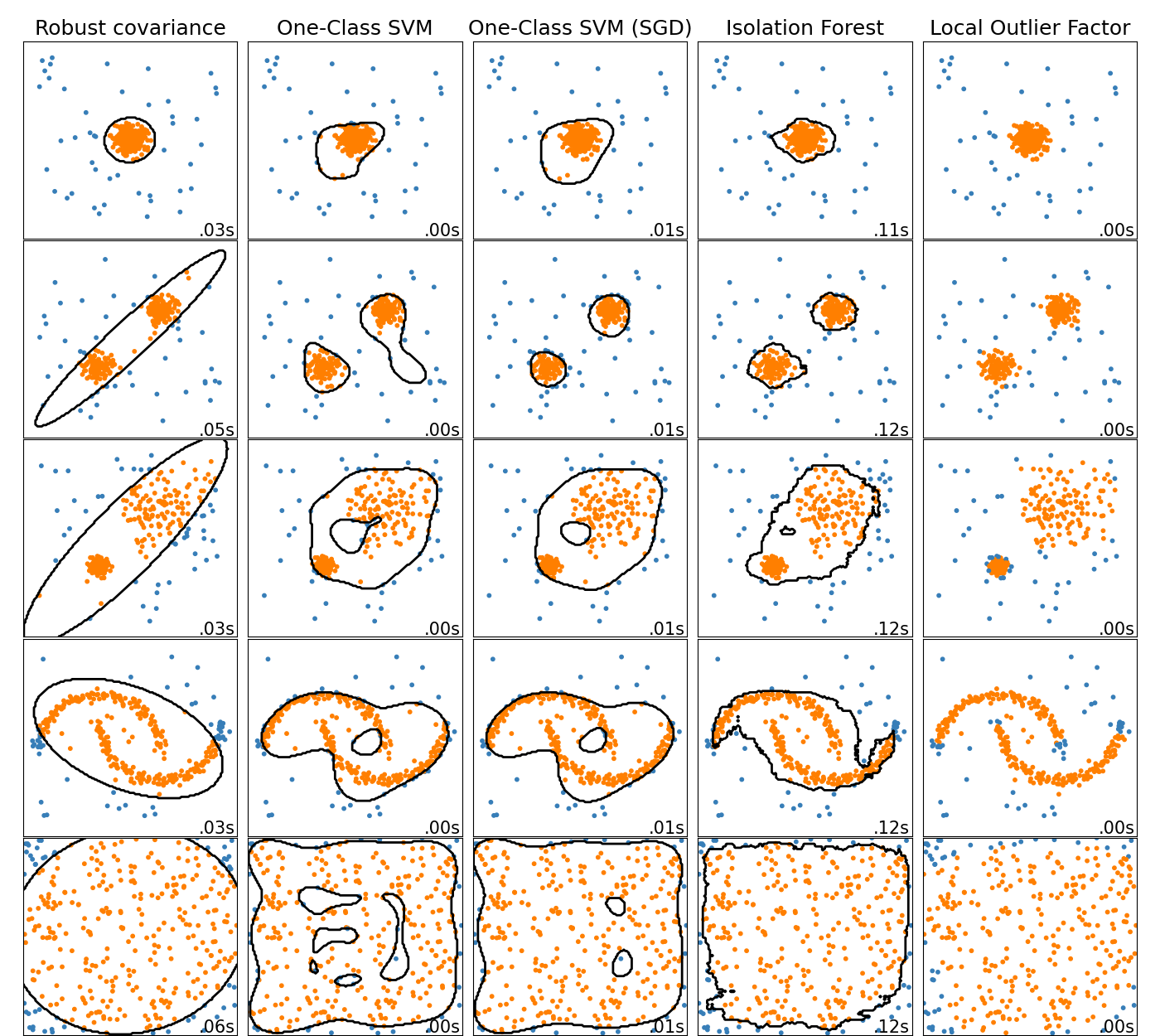
Online One-Class SVM¶
The new class SGDOneClassSVM implements an online
linear version of the One-Class SVM using a stochastic gradient descent.
Combined with kernel approximation techniques,
SGDOneClassSVM can be used to approximate the solution
of a kernelized One-Class SVM, implemented in OneClassSVM, with
a fit time complexity linear in the number of samples. Note that the
complexity of a kernelized One-Class SVM is at best quadratic in the number
of samples. SGDOneClassSVM is thus well suited for
datasets with a large number of training samples (> 10,000) for which the SGD
variant can be several orders of magnitude faster. Please check this
example to see how
it’s used, and the User Guide for more
details.
Histogram-based Gradient Boosting Models are now stable¶
HistGradientBoostingRegressor and
HistGradientBoostingClassifier are no longer experimental
and can simply be imported and used as:
from sklearn.ensemble import HistGradientBoostingClassifier
New documentation improvements¶
This release includes many documentation improvements. Out of over 2100 merged pull requests, about 800 of them are improvements to our documentation.
Total running time of the script: (0 minutes 0.016 seconds)