Note
Click here to download the full example code or to run this example in your browser via Binder
Out-of-core classification of text documents¶
This is an example showing how scikit-learn can be used for classification using an out-of-core approach: learning from data that doesn’t fit into main memory. We make use of an online classifier, i.e., one that supports the partial_fit method, that will be fed with batches of examples. To guarantee that the features space remains the same over time we leverage a HashingVectorizer that will project each example into the same feature space. This is especially useful in the case of text classification where new features (words) may appear in each batch.
# Authors: Eustache Diemert <eustache@diemert.fr>
# @FedericoV <https://github.com/FedericoV/>
# License: BSD 3 clause
from glob import glob
import itertools
import os.path
import re
import tarfile
import time
import sys
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rcParams
from html.parser import HTMLParser
from urllib.request import urlretrieve
from sklearn.datasets import get_data_home
from sklearn.feature_extraction.text import HashingVectorizer
from sklearn.linear_model import SGDClassifier
from sklearn.linear_model import PassiveAggressiveClassifier
from sklearn.linear_model import Perceptron
from sklearn.naive_bayes import MultinomialNB
def _not_in_sphinx():
# Hack to detect whether we are running by the sphinx builder
return '__file__' in globals()
Main¶
Create the vectorizer and limit the number of features to a reasonable maximum
vectorizer = HashingVectorizer(decode_error='ignore', n_features=2 ** 18,
alternate_sign=False)
# Iterator over parsed Reuters SGML files.
data_stream = stream_reuters_documents()
# We learn a binary classification between the "acq" class and all the others.
# "acq" was chosen as it is more or less evenly distributed in the Reuters
# files. For other datasets, one should take care of creating a test set with
# a realistic portion of positive instances.
all_classes = np.array([0, 1])
positive_class = 'acq'
# Here are some classifiers that support the `partial_fit` method
partial_fit_classifiers = {
'SGD': SGDClassifier(max_iter=5),
'Perceptron': Perceptron(),
'NB Multinomial': MultinomialNB(alpha=0.01),
'Passive-Aggressive': PassiveAggressiveClassifier(),
}
def get_minibatch(doc_iter, size, pos_class=positive_class):
"""Extract a minibatch of examples, return a tuple X_text, y.
Note: size is before excluding invalid docs with no topics assigned.
"""
data = [('{title}\n\n{body}'.format(**doc), pos_class in doc['topics'])
for doc in itertools.islice(doc_iter, size)
if doc['topics']]
if not len(data):
return np.asarray([], dtype=int), np.asarray([], dtype=int)
X_text, y = zip(*data)
return X_text, np.asarray(y, dtype=int)
def iter_minibatches(doc_iter, minibatch_size):
"""Generator of minibatches."""
X_text, y = get_minibatch(doc_iter, minibatch_size)
while len(X_text):
yield X_text, y
X_text, y = get_minibatch(doc_iter, minibatch_size)
# test data statistics
test_stats = {'n_test': 0, 'n_test_pos': 0}
# First we hold out a number of examples to estimate accuracy
n_test_documents = 1000
tick = time.time()
X_test_text, y_test = get_minibatch(data_stream, 1000)
parsing_time = time.time() - tick
tick = time.time()
X_test = vectorizer.transform(X_test_text)
vectorizing_time = time.time() - tick
test_stats['n_test'] += len(y_test)
test_stats['n_test_pos'] += sum(y_test)
print("Test set is %d documents (%d positive)" % (len(y_test), sum(y_test)))
def progress(cls_name, stats):
"""Report progress information, return a string."""
duration = time.time() - stats['t0']
s = "%20s classifier : \t" % cls_name
s += "%(n_train)6d train docs (%(n_train_pos)6d positive) " % stats
s += "%(n_test)6d test docs (%(n_test_pos)6d positive) " % test_stats
s += "accuracy: %(accuracy).3f " % stats
s += "in %.2fs (%5d docs/s)" % (duration, stats['n_train'] / duration)
return s
cls_stats = {}
for cls_name in partial_fit_classifiers:
stats = {'n_train': 0, 'n_train_pos': 0,
'accuracy': 0.0, 'accuracy_history': [(0, 0)], 't0': time.time(),
'runtime_history': [(0, 0)], 'total_fit_time': 0.0}
cls_stats[cls_name] = stats
get_minibatch(data_stream, n_test_documents)
# Discard test set
# We will feed the classifier with mini-batches of 1000 documents; this means
# we have at most 1000 docs in memory at any time. The smaller the document
# batch, the bigger the relative overhead of the partial fit methods.
minibatch_size = 1000
# Create the data_stream that parses Reuters SGML files and iterates on
# documents as a stream.
minibatch_iterators = iter_minibatches(data_stream, minibatch_size)
total_vect_time = 0.0
# Main loop : iterate on mini-batches of examples
for i, (X_train_text, y_train) in enumerate(minibatch_iterators):
tick = time.time()
X_train = vectorizer.transform(X_train_text)
total_vect_time += time.time() - tick
for cls_name, cls in partial_fit_classifiers.items():
tick = time.time()
# update estimator with examples in the current mini-batch
cls.partial_fit(X_train, y_train, classes=all_classes)
# accumulate test accuracy stats
cls_stats[cls_name]['total_fit_time'] += time.time() - tick
cls_stats[cls_name]['n_train'] += X_train.shape[0]
cls_stats[cls_name]['n_train_pos'] += sum(y_train)
tick = time.time()
cls_stats[cls_name]['accuracy'] = cls.score(X_test, y_test)
cls_stats[cls_name]['prediction_time'] = time.time() - tick
acc_history = (cls_stats[cls_name]['accuracy'],
cls_stats[cls_name]['n_train'])
cls_stats[cls_name]['accuracy_history'].append(acc_history)
run_history = (cls_stats[cls_name]['accuracy'],
total_vect_time + cls_stats[cls_name]['total_fit_time'])
cls_stats[cls_name]['runtime_history'].append(run_history)
if i % 3 == 0:
print(progress(cls_name, cls_stats[cls_name]))
if i % 3 == 0:
print('\n')
Out:
Test set is 973 documents (125 positive)
SGD classifier : 965 train docs ( 134 positive) 973 test docs ( 125 positive) accuracy: 0.921 in 0.68s ( 1419 docs/s)
Perceptron classifier : 965 train docs ( 134 positive) 973 test docs ( 125 positive) accuracy: 0.872 in 0.69s ( 1407 docs/s)
NB Multinomial classifier : 965 train docs ( 134 positive) 973 test docs ( 125 positive) accuracy: 0.874 in 0.71s ( 1355 docs/s)
Passive-Aggressive classifier : 965 train docs ( 134 positive) 973 test docs ( 125 positive) accuracy: 0.920 in 0.71s ( 1350 docs/s)
SGD classifier : 3790 train docs ( 506 positive) 973 test docs ( 125 positive) accuracy: 0.963 in 1.96s ( 1932 docs/s)
Perceptron classifier : 3790 train docs ( 506 positive) 973 test docs ( 125 positive) accuracy: 0.949 in 1.96s ( 1929 docs/s)
NB Multinomial classifier : 3790 train docs ( 506 positive) 973 test docs ( 125 positive) accuracy: 0.884 in 1.98s ( 1912 docs/s)
Passive-Aggressive classifier : 3790 train docs ( 506 positive) 973 test docs ( 125 positive) accuracy: 0.947 in 1.98s ( 1910 docs/s)
SGD classifier : 6523 train docs ( 916 positive) 973 test docs ( 125 positive) accuracy: 0.950 in 3.21s ( 2031 docs/s)
Perceptron classifier : 6523 train docs ( 916 positive) 973 test docs ( 125 positive) accuracy: 0.923 in 3.21s ( 2030 docs/s)
NB Multinomial classifier : 6523 train docs ( 916 positive) 973 test docs ( 125 positive) accuracy: 0.909 in 3.23s ( 2019 docs/s)
Passive-Aggressive classifier : 6523 train docs ( 916 positive) 973 test docs ( 125 positive) accuracy: 0.953 in 3.23s ( 2017 docs/s)
SGD classifier : 9434 train docs ( 1242 positive) 973 test docs ( 125 positive) accuracy: 0.927 in 4.48s ( 2107 docs/s)
Perceptron classifier : 9434 train docs ( 1242 positive) 973 test docs ( 125 positive) accuracy: 0.947 in 4.48s ( 2106 docs/s)
NB Multinomial classifier : 9434 train docs ( 1242 positive) 973 test docs ( 125 positive) accuracy: 0.918 in 4.50s ( 2098 docs/s)
Passive-Aggressive classifier : 9434 train docs ( 1242 positive) 973 test docs ( 125 positive) accuracy: 0.956 in 4.50s ( 2096 docs/s)
SGD classifier : 11845 train docs ( 1468 positive) 973 test docs ( 125 positive) accuracy: 0.949 in 5.75s ( 2061 docs/s)
Perceptron classifier : 11845 train docs ( 1468 positive) 973 test docs ( 125 positive) accuracy: 0.942 in 5.75s ( 2060 docs/s)
NB Multinomial classifier : 11845 train docs ( 1468 positive) 973 test docs ( 125 positive) accuracy: 0.922 in 5.77s ( 2054 docs/s)
Passive-Aggressive classifier : 11845 train docs ( 1468 positive) 973 test docs ( 125 positive) accuracy: 0.942 in 5.77s ( 2053 docs/s)
SGD classifier : 14770 train docs ( 1856 positive) 973 test docs ( 125 positive) accuracy: 0.959 in 7.10s ( 2079 docs/s)
Perceptron classifier : 14770 train docs ( 1856 positive) 973 test docs ( 125 positive) accuracy: 0.956 in 7.11s ( 2078 docs/s)
NB Multinomial classifier : 14770 train docs ( 1856 positive) 973 test docs ( 125 positive) accuracy: 0.925 in 7.12s ( 2073 docs/s)
Passive-Aggressive classifier : 14770 train docs ( 1856 positive) 973 test docs ( 125 positive) accuracy: 0.957 in 7.13s ( 2072 docs/s)
SGD classifier : 17723 train docs ( 2218 positive) 973 test docs ( 125 positive) accuracy: 0.959 in 8.47s ( 2093 docs/s)
Perceptron classifier : 17723 train docs ( 2218 positive) 973 test docs ( 125 positive) accuracy: 0.938 in 8.47s ( 2092 docs/s)
NB Multinomial classifier : 17723 train docs ( 2218 positive) 973 test docs ( 125 positive) accuracy: 0.925 in 8.49s ( 2087 docs/s)
Passive-Aggressive classifier : 17723 train docs ( 2218 positive) 973 test docs ( 125 positive) accuracy: 0.962 in 8.49s ( 2087 docs/s)
Plot results¶
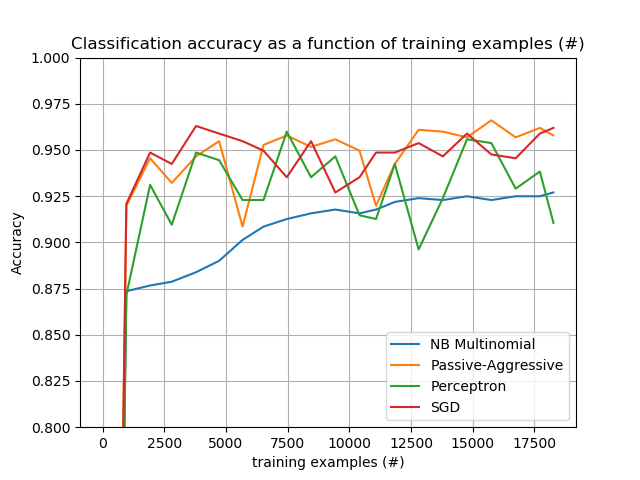
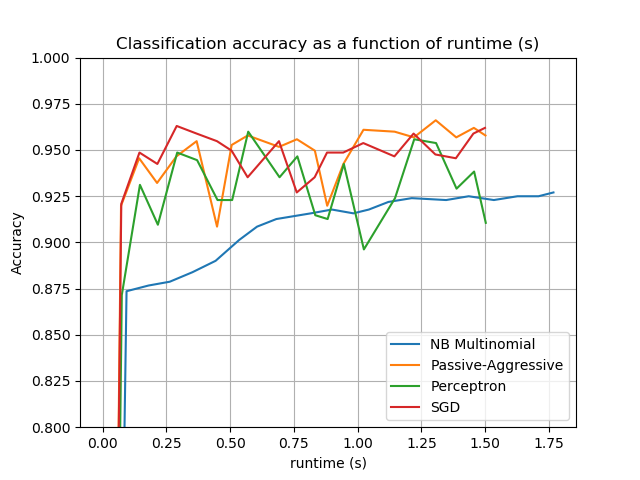
The plot represents the learning curve of the classifier: the evolution of classification accuracy over the course of the mini-batches. Accuracy is measured on the first 1000 samples, held out as a validation set.
To limit the memory consumption, we queue examples up to a fixed amount before feeding them to the learner.
def plot_accuracy(x, y, x_legend):
"""Plot accuracy as a function of x."""
x = np.array(x)
y = np.array(y)
plt.title('Classification accuracy as a function of %s' % x_legend)
plt.xlabel('%s' % x_legend)
plt.ylabel('Accuracy')
plt.grid(True)
plt.plot(x, y)
rcParams['legend.fontsize'] = 10
cls_names = list(sorted(cls_stats.keys()))
# Plot accuracy evolution
plt.figure()
for _, stats in sorted(cls_stats.items()):
# Plot accuracy evolution with #examples
accuracy, n_examples = zip(*stats['accuracy_history'])
plot_accuracy(n_examples, accuracy, "training examples (#)")
ax = plt.gca()
ax.set_ylim((0.8, 1))
plt.legend(cls_names, loc='best')
plt.figure()
for _, stats in sorted(cls_stats.items()):
# Plot accuracy evolution with runtime
accuracy, runtime = zip(*stats['runtime_history'])
plot_accuracy(runtime, accuracy, 'runtime (s)')
ax = plt.gca()
ax.set_ylim((0.8, 1))
plt.legend(cls_names, loc='best')
# Plot fitting times
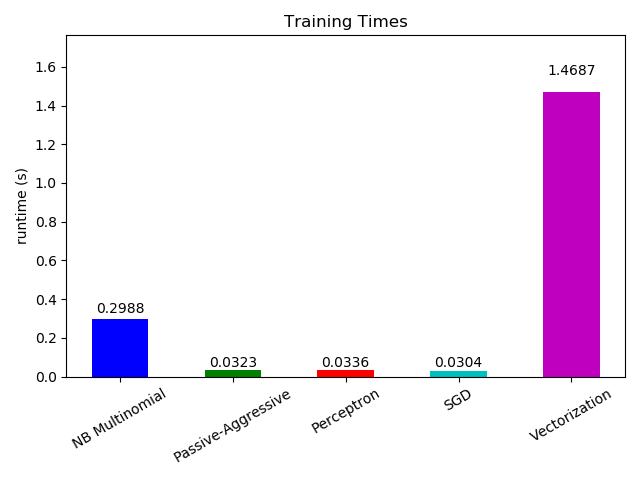
plt.figure()
fig = plt.gcf()
cls_runtime = [stats['total_fit_time']
for cls_name, stats in sorted(cls_stats.items())]
cls_runtime.append(total_vect_time)
cls_names.append('Vectorization')
bar_colors = ['b', 'g', 'r', 'c', 'm', 'y']
ax = plt.subplot(111)
rectangles = plt.bar(range(len(cls_names)), cls_runtime, width=0.5,
color=bar_colors)
ax.set_xticks(np.linspace(0, len(cls_names) - 1, len(cls_names)))
ax.set_xticklabels(cls_names, fontsize=10)
ymax = max(cls_runtime) * 1.2
ax.set_ylim((0, ymax))
ax.set_ylabel('runtime (s)')
ax.set_title('Training Times')
def autolabel(rectangles):
"""attach some text vi autolabel on rectangles."""
for rect in rectangles:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width() / 2.,
1.05 * height, '%.4f' % height,
ha='center', va='bottom')
plt.setp(plt.xticks()[1], rotation=30)
autolabel(rectangles)
plt.tight_layout()
plt.show()
# Plot prediction times
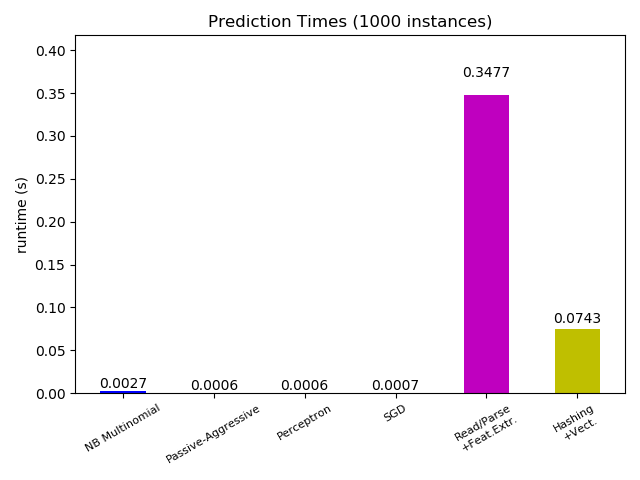
plt.figure()
cls_runtime = []
cls_names = list(sorted(cls_stats.keys()))
for cls_name, stats in sorted(cls_stats.items()):
cls_runtime.append(stats['prediction_time'])
cls_runtime.append(parsing_time)
cls_names.append('Read/Parse\n+Feat.Extr.')
cls_runtime.append(vectorizing_time)
cls_names.append('Hashing\n+Vect.')
ax = plt.subplot(111)
rectangles = plt.bar(range(len(cls_names)), cls_runtime, width=0.5,
color=bar_colors)
ax.set_xticks(np.linspace(0, len(cls_names) - 1, len(cls_names)))
ax.set_xticklabels(cls_names, fontsize=8)
plt.setp(plt.xticks()[1], rotation=30)
ymax = max(cls_runtime) * 1.2
ax.set_ylim((0, ymax))
ax.set_ylabel('runtime (s)')
ax.set_title('Prediction Times (%d instances)' % n_test_documents)
autolabel(rectangles)
plt.tight_layout()
plt.show()
Total running time of the script: ( 0 minutes 10.253 seconds)
Estimated memory usage: 8 MB