Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
Theil-Sen Regression¶
Computes a Theil-Sen Regression on a synthetic dataset.
See Theil-Sen estimator: generalized-median-based estimator for more information on the regressor.
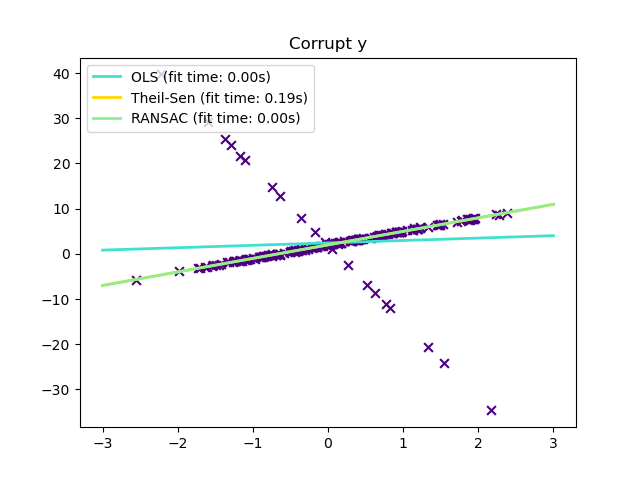
Compared to the OLS (ordinary least squares) estimator, the Theil-Sen estimator is robust against outliers. It has a breakdown point of about 29.3% in case of a simple linear regression which means that it can tolerate arbitrary corrupted data (outliers) of up to 29.3% in the two-dimensional case.
The estimation of the model is done by calculating the slopes and intercepts of a subpopulation of all possible combinations of p subsample points. If an intercept is fitted, p must be greater than or equal to n_features + 1. The final slope and intercept is then defined as the spatial median of these slopes and intercepts.
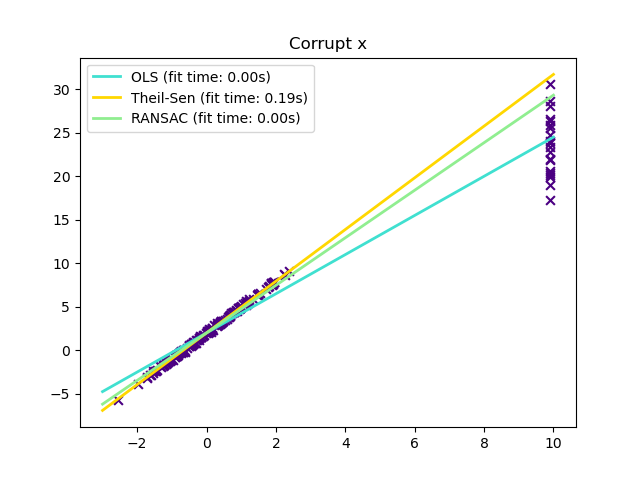
In certain cases Theil-Sen performs better than RANSAC which is also a robust method. This is illustrated in the
second example below where outliers with respect to the x-axis perturb RANSAC.
Tuning the residual_threshold parameter of RANSAC remedies this but in
general a priori knowledge about the data and the nature of the outliers is
needed.
Due to the computational complexity of Theil-Sen it is recommended to use it
only for small problems in terms of number of samples and features. For larger
problems the max_subpopulation parameter restricts the magnitude of all
possible combinations of p subsample points to a randomly chosen subset and
therefore also limits the runtime. Therefore, Theil-Sen is applicable to larger
problems with the drawback of losing some of its mathematical properties since
it then works on a random subset.
# Author: Florian Wilhelm -- <florian.wilhelm@gmail.com>
# License: BSD 3 clause
import time
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import LinearRegression, RANSACRegressor, TheilSenRegressor
estimators = [
("OLS", LinearRegression()),
("Theil-Sen", TheilSenRegressor(random_state=42)),
("RANSAC", RANSACRegressor(random_state=42)),
]
colors = {"OLS": "turquoise", "Theil-Sen": "gold", "RANSAC": "lightgreen"}
lw = 2
Outliers only in the y direction¶
np.random.seed(0)
n_samples = 200
# Linear model y = 3*x + N(2, 0.1**2)
x = np.random.randn(n_samples)
w = 3.0
c = 2.0
noise = 0.1 * np.random.randn(n_samples)
y = w * x + c + noise
# 10% outliers
y[-20:] += -20 * x[-20:]
X = x[:, np.newaxis]
plt.scatter(x, y, color="indigo", marker="x", s=40)
line_x = np.array([-3, 3])
for name, estimator in estimators:
t0 = time.time()
estimator.fit(X, y)
elapsed_time = time.time() - t0
y_pred = estimator.predict(line_x.reshape(2, 1))
plt.plot(
line_x,
y_pred,
color=colors[name],
linewidth=lw,
label="%s (fit time: %.2fs)" % (name, elapsed_time),
)
plt.axis("tight")
plt.legend(loc="upper left")
_ = plt.title("Corrupt y")
Outliers in the X direction¶
np.random.seed(0)
# Linear model y = 3*x + N(2, 0.1**2)
x = np.random.randn(n_samples)
noise = 0.1 * np.random.randn(n_samples)
y = 3 * x + 2 + noise
# 10% outliers
x[-20:] = 9.9
y[-20:] += 22
X = x[:, np.newaxis]
plt.figure()
plt.scatter(x, y, color="indigo", marker="x", s=40)
line_x = np.array([-3, 10])
for name, estimator in estimators:
t0 = time.time()
estimator.fit(X, y)
elapsed_time = time.time() - t0
y_pred = estimator.predict(line_x.reshape(2, 1))
plt.plot(
line_x,
y_pred,
color=colors[name],
linewidth=lw,
label="%s (fit time: %.2fs)" % (name, elapsed_time),
)
plt.axis("tight")
plt.legend(loc="upper left")
plt.title("Corrupt x")
plt.show()
Total running time of the script: (0 minutes 0.546 seconds)
Related examples
Compare the effect of different scalers on data with outliers
Comparing anomaly detection algorithms for outlier detection on toy datasets