Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
L1-based models for Sparse Signals¶
The present example compares three l1-based regression models on a synthetic signal obtained from sparse and correlated features that are further corrupted with additive gaussian noise:
It is known that the Lasso estimates turn to be close to the model selection estimates when the data dimensions grow, given that the irrelevant variables are not too correlated with the relevant ones. In the presence of correlated features, Lasso itself cannot select the correct sparsity pattern [1].
Here we compare the performance of the three models in terms of the \(R^2\) score, the fitting time and the sparsity of the estimated coefficients when compared with the ground-truth.
# Author: Arturo Amor <david-arturo.amor-quiroz@inria.fr>
Generate synthetic dataset¶
We generate a dataset where the number of samples is lower than the total number of features. This leads to an underdetermined system, i.e. the solution is not unique, and thus we cannot apply an Ordinary Least Squares by itself. Regularization introduces a penalty term to the objective function, which modifies the optimization problem and can help alleviate the underdetermined nature of the system.
The target y is a linear combination with alternating signs of sinusoidal
signals. Only the 10 lowest out of the 100 frequencies in X are used to
generate y, while the rest of the features are not informative. This results
in a high dimensional sparse feature space, where some degree of
l1-penalization is necessary.
import numpy as np
rng = np.random.RandomState(0)
n_samples, n_features, n_informative = 50, 100, 10
time_step = np.linspace(-2, 2, n_samples)
freqs = 2 * np.pi * np.sort(rng.rand(n_features)) / 0.01
X = np.zeros((n_samples, n_features))
for i in range(n_features):
X[:, i] = np.sin(freqs[i] * time_step)
idx = np.arange(n_features)
true_coef = (-1) ** idx * np.exp(-idx / 10)
true_coef[n_informative:] = 0 # sparsify coef
y = np.dot(X, true_coef)
Some of the informative features have close frequencies to induce (anti-)correlations.
freqs[:n_informative]
array([ 2.9502547 , 11.8059798 , 12.63394388, 12.70359377, 24.62241605,
37.84077985, 40.30506066, 44.63327171, 54.74495357, 59.02456369])
A random phase is introduced using numpy.random.random_sample
and some gaussian noise (implemented by numpy.random.normal)
is added to both the features and the target.
for i in range(n_features):
X[:, i] = np.sin(freqs[i] * time_step + 2 * (rng.random_sample() - 0.5))
X[:, i] += 0.2 * rng.normal(0, 1, n_samples)
y += 0.2 * rng.normal(0, 1, n_samples)
Such sparse, noisy and correlated features can be obtained, for instance, from sensor nodes monitoring some environmental variables, as they typically register similar values depending on their positions (spatial correlations). We can visualize the target.
import matplotlib.pyplot as plt
plt.plot(time_step, y)
plt.ylabel("target signal")
plt.xlabel("time")
_ = plt.title("Superposition of sinusoidal signals")
We split the data into train and test sets for simplicity. In practice one
should use a TimeSeriesSplit
cross-validation to estimate the variance of the test score. Here we set
shuffle="False" as we must not use training data that succeed the testing
data when dealing with data that have a temporal relationship.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5, shuffle=False)
In the following, we compute the performance of three l1-based models in terms of the goodness of fit \(R^2\) score and the fitting time. Then we make a plot to compare the sparsity of the estimated coefficients with respect to the ground-truth coefficients and finally we analyze the previous results.
Lasso¶
In this example, we demo a Lasso with a fixed
value of the regularization parameter alpha. In practice, the optimal
parameter alpha should be selected by passing a
TimeSeriesSplit cross-validation strategy to a
LassoCV. To keep the example simple and fast to
execute, we directly set the optimal value for alpha here.
from time import time
from sklearn.linear_model import Lasso
from sklearn.metrics import r2_score
t0 = time()
lasso = Lasso(alpha=0.14).fit(X_train, y_train)
print(f"Lasso fit done in {(time() - t0):.3f}s")
y_pred_lasso = lasso.predict(X_test)
r2_score_lasso = r2_score(y_test, y_pred_lasso)
print(f"Lasso r^2 on test data : {r2_score_lasso:.3f}")
Lasso fit done in 0.001s
Lasso r^2 on test data : 0.480
Automatic Relevance Determination (ARD)¶
An ARD regression is the bayesian version of the Lasso. It can produce
interval estimates for all of the parameters, including the error variance, if
required. It is a suitable option when the signals have gaussian noise. See
the example Comparing Linear Bayesian Regressors for a
comparison of ARDRegression and
BayesianRidge regressors.
from sklearn.linear_model import ARDRegression
t0 = time()
ard = ARDRegression().fit(X_train, y_train)
print(f"ARD fit done in {(time() - t0):.3f}s")
y_pred_ard = ard.predict(X_test)
r2_score_ard = r2_score(y_test, y_pred_ard)
print(f"ARD r^2 on test data : {r2_score_ard:.3f}")
ARD fit done in 0.124s
ARD r^2 on test data : 0.542
ElasticNet¶
ElasticNet is a middle ground between
Lasso and Ridge,
as it combines a L1 and a L2-penalty. The amount of regularization is
controlled by the two hyperparameters l1_ratio and alpha. For l1_ratio =
0 the penalty is pure L2 and the model is equivalent to a
Ridge. Similarly, l1_ratio = 1 is a pure L1
penalty and the model is equivalent to a Lasso.
For 0 < l1_ratio < 1, the penalty is a combination of L1 and L2.
As done before, we train the model with fix values for alpha and l1_ratio.
To select their optimal value we used an
ElasticNetCV, not shown here to keep the
example simple.
from sklearn.linear_model import ElasticNet
t0 = time()
enet = ElasticNet(alpha=0.08, l1_ratio=0.5).fit(X_train, y_train)
print(f"ElasticNet fit done in {(time() - t0):.3f}s")
y_pred_enet = enet.predict(X_test)
r2_score_enet = r2_score(y_test, y_pred_enet)
print(f"ElasticNet r^2 on test data : {r2_score_enet:.3f}")
ElasticNet fit done in 0.002s
ElasticNet r^2 on test data : 0.636
Plot and analysis of the results¶
In this section, we use a heatmap to visualize the sparsity of the true and estimated coefficients of the respective linear models.
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from matplotlib.colors import SymLogNorm
df = pd.DataFrame(
{
"True coefficients": true_coef,
"Lasso": lasso.coef_,
"ARDRegression": ard.coef_,
"ElasticNet": enet.coef_,
}
)
plt.figure(figsize=(10, 6))
ax = sns.heatmap(
df.T,
norm=SymLogNorm(linthresh=10e-4, vmin=-1, vmax=1),
cbar_kws={"label": "coefficients' values"},
cmap="seismic_r",
)
plt.ylabel("linear model")
plt.xlabel("coefficients")
plt.title(
f"Models' coefficients\nLasso $R^2$: {r2_score_lasso:.3f}, "
f"ARD $R^2$: {r2_score_ard:.3f}, "
f"ElasticNet $R^2$: {r2_score_enet:.3f}"
)
plt.tight_layout()
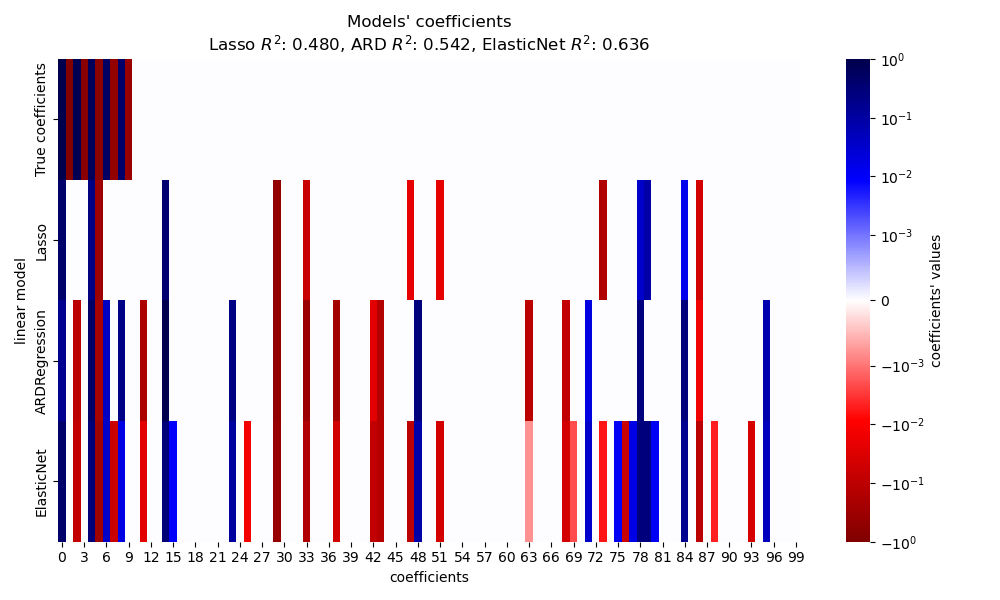
In the present example ElasticNet yields the
best score and captures the most of the predictive features, yet still fails
at finding all the true components. Notice that both
ElasticNet and
ARDRegression result in a less sparse model
than a Lasso.
Conclusions¶
Lasso is known to recover sparse data
effectively but does not perform well with highly correlated features. Indeed,
if several correlated features contribute to the target,
Lasso would end up selecting a single one of
them. In the case of sparse yet non-correlated features, a
Lasso model would be more suitable.
ElasticNet introduces some sparsity on the
coefficients and shrinks their values to zero. Thus, in the presence of
correlated features that contribute to the target, the model is still able to
reduce their weights without setting them exactly to zero. This results in a
less sparse model than a pure Lasso and may
capture non-predictive features as well.
ARDRegression is better when handling gaussian
noise, but is still unable to handle correlated features and requires a larger
amount of time due to fitting a prior.
References¶
Total running time of the script: (0 minutes 0.625 seconds)