sklearn.cluster.DBSCAN¶
- class sklearn.cluster.DBSCAN(eps=0.5, min_samples=5, metric='euclidean', algorithm='auto', leaf_size=30, p=None, random_state=None)¶
Perform DBSCAN clustering from vector array or distance matrix.
DBSCAN - Density-Based Spatial Clustering of Applications with Noise. Finds core samples of high density and expands clusters from them. Good for data which contains clusters of similar density.
Parameters: eps : float, optional
The maximum distance between two samples for them to be considered as in the same neighborhood.
min_samples : int, optional
The number of samples in a neighborhood for a point to be considered as a core point.
metric : string, or callable
The metric to use when calculating distance between instances in a feature array. If metric is a string or callable, it must be one of the options allowed by metrics.pairwise.calculate_distance for its metric parameter. If metric is “precomputed”, X is assumed to be a distance matrix and must be square.
random_state : numpy.RandomState, optional
The generator used to initialize the centers. Defaults to numpy.random.
Attributes: `core_sample_indices_` : array, shape = [n_core_samples]
Indices of core samples.
`components_` : array, shape = [n_core_samples, n_features]
Copy of each core sample found by training.
`labels_` : array, shape = [n_samples]
Cluster labels for each point in the dataset given to fit(). Noisy samples are given the label -1.
Notes
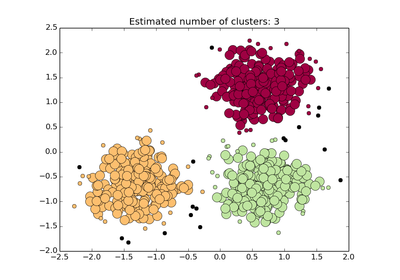
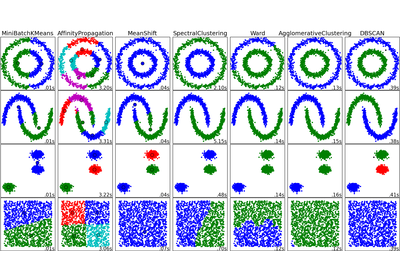
See examples/plot_dbscan.py for an example.
References
Ester, M., H. P. Kriegel, J. Sander, and X. Xu, “A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise”. In: Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining, Portland, OR, AAAI Press, pp. 226-231. 1996
Methods
fit(X) Perform DBSCAN clustering from features or distance matrix. fit_predict(X[, y]) Performs clustering on X and returns cluster labels. get_params([deep]) Get parameters for this estimator. set_params(**params) Set the parameters of this estimator. - __init__(eps=0.5, min_samples=5, metric='euclidean', algorithm='auto', leaf_size=30, p=None, random_state=None)¶
- fit(X)¶
Perform DBSCAN clustering from features or distance matrix.
Parameters: X: array [n_samples, n_samples] or [n_samples, n_features] :
Array of distances between samples, or a feature array. The array is treated as a feature array unless the metric is given as ‘precomputed’.
params: dict :
Overwrite keywords from __init__.
- fit_predict(X, y=None)¶
Performs clustering on X and returns cluster labels.
Parameters: X : ndarray, shape (n_samples, n_features)
Input data.
Returns: y : ndarray, shape (n_samples,)
cluster labels
- get_params(deep=True)¶
Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The former have parameters of the form <component>__<parameter> so that it’s possible to update each component of a nested object.
Returns: self :