sklearn.impute.SimpleImputer¶
- class sklearn.impute.SimpleImputer(*, missing_values=nan, strategy='mean', fill_value=None, copy=True, add_indicator=False, keep_empty_features=False)[source]¶
Univariate imputer for completing missing values with simple strategies.
Replace missing values using a descriptive statistic (e.g. mean, median, or most frequent) along each column, or using a constant value.
Read more in the User Guide.
New in version 0.20:
SimpleImputerreplaces the previoussklearn.preprocessing.Imputerestimator which is now removed.- Parameters:
- missing_valuesint, float, str, np.nan, None or pandas.NA, default=np.nan
The placeholder for the missing values. All occurrences of
missing_valueswill be imputed. For pandas’ dataframes with nullable integer dtypes with missing values,missing_valuescan be set to eithernp.nanorpd.NA.- strategystr, default=’mean’
The imputation strategy.
If “mean”, then replace missing values using the mean along each column. Can only be used with numeric data.
If “median”, then replace missing values using the median along each column. Can only be used with numeric data.
If “most_frequent”, then replace missing using the most frequent value along each column. Can be used with strings or numeric data. If there is more than one such value, only the smallest is returned.
If “constant”, then replace missing values with fill_value. Can be used with strings or numeric data.
New in version 0.20: strategy=”constant” for fixed value imputation.
- fill_valuestr or numerical value, default=None
When strategy == “constant”,
fill_valueis used to replace all occurrences of missing_values. For string or object data types,fill_valuemust be a string. IfNone,fill_valuewill be 0 when imputing numerical data and “missing_value” for strings or object data types.- copybool, default=True
If True, a copy of X will be created. If False, imputation will be done in-place whenever possible. Note that, in the following cases, a new copy will always be made, even if
copy=False:If
Xis not an array of floating values;If
Xis encoded as a CSR matrix;If
add_indicator=True.
- add_indicatorbool, default=False
If True, a
MissingIndicatortransform will stack onto output of the imputer’s transform. This allows a predictive estimator to account for missingness despite imputation. If a feature has no missing values at fit/train time, the feature won’t appear on the missing indicator even if there are missing values at transform/test time.- keep_empty_featuresbool, default=False
If True, features that consist exclusively of missing values when
fitis called are returned in results whentransformis called. The imputed value is always0except whenstrategy="constant"in which casefill_valuewill be used instead.New in version 1.2.
- Attributes:
- statistics_array of shape (n_features,)
The imputation fill value for each feature. Computing statistics can result in
np.nanvalues. Duringtransform, features corresponding tonp.nanstatistics will be discarded.- indicator_
MissingIndicator Indicator used to add binary indicators for missing values.
Noneifadd_indicator=False.- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
IterativeImputerMultivariate imputer that estimates values to impute for each feature with missing values from all the others.
KNNImputerMultivariate imputer that estimates missing features using nearest samples.
Notes
Columns which only contained missing values at
fitare discarded upontransformif strategy is not"constant".In a prediction context, simple imputation usually performs poorly when associated with a weak learner. However, with a powerful learner, it can lead to as good or better performance than complex imputation such as
IterativeImputerorKNNImputer.Examples
>>> import numpy as np >>> from sklearn.impute import SimpleImputer >>> imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean') >>> imp_mean.fit([[7, 2, 3], [4, np.nan, 6], [10, 5, 9]]) SimpleImputer() >>> X = [[np.nan, 2, 3], [4, np.nan, 6], [10, np.nan, 9]] >>> print(imp_mean.transform(X)) [[ 7. 2. 3. ] [ 4. 3.5 6. ] [10. 3.5 9. ]]
For a more detailed example see Imputing missing values before building an estimator.
Methods
fit(X[, y])Fit the imputer on
X.fit_transform(X[, y])Fit to data, then transform it.
get_feature_names_out([input_features])Get output feature names for transformation.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
Convert the data back to the original representation.
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Impute all missing values in
X.- fit(X, y=None)[source]¶
Fit the imputer on
X.- Parameters:
- X{array-like, sparse matrix}, shape (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.- yIgnored
Not used, present here for API consistency by convention.
- Returns:
- selfobject
Fitted estimator.
- fit_transform(X, y=None, **fit_params)[source]¶
Fit to data, then transform it.
Fits transformer to
Xandywith optional parametersfit_paramsand returns a transformed version ofX.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs), default=None
Target values (None for unsupervised transformations).
- **fit_paramsdict
Additional fit parameters.
- Returns:
- X_newndarray array of shape (n_samples, n_features_new)
Transformed array.
- get_feature_names_out(input_features=None)[source]¶
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names.
- get_metadata_routing()[source]¶
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- inverse_transform(X)[source]¶
Convert the data back to the original representation.
Inverts the
transformoperation performed on an array. This operation can only be performed afterSimpleImputeris instantiated withadd_indicator=True.Note that
inverse_transformcan only invert the transform in features that have binary indicators for missing values. If a feature has no missing values atfittime, the feature won’t have a binary indicator, and the imputation done attransformtime won’t be inverted.New in version 0.24.
- Parameters:
- Xarray-like of shape (n_samples, n_features + n_features_missing_indicator)
The imputed data to be reverted to original data. It has to be an augmented array of imputed data and the missing indicator mask.
- Returns:
- X_originalndarray of shape (n_samples, n_features)
The original
Xwith missing values as it was prior to imputation.
- set_output(*, transform=None)[source]¶
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame outputNone: Transform configuration is unchanged
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using sklearn.impute.SimpleImputer¶
Permutation Importance vs Random Forest Feature Importance (MDI)
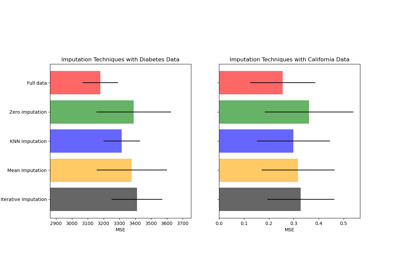
Imputing missing values before building an estimator
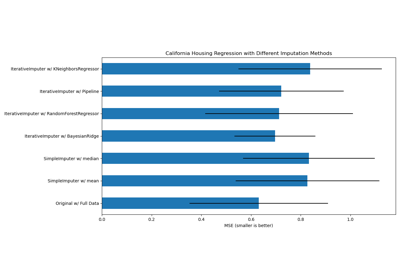
Imputing missing values with variants of IterativeImputer