sklearn.preprocessing.OrdinalEncoder¶
- class sklearn.preprocessing.OrdinalEncoder(*, categories='auto', dtype=<class 'numpy.float64'>, handle_unknown='error', unknown_value=None, encoded_missing_value=nan)[source]¶
Encode categorical features as an integer array.
The input to this transformer should be an array-like of integers or strings, denoting the values taken on by categorical (discrete) features. The features are converted to ordinal integers. This results in a single column of integers (0 to n_categories - 1) per feature.
Read more in the User Guide.
New in version 0.20.
- Parameters:
- categories‘auto’ or a list of array-like, default=’auto’
Categories (unique values) per feature:
‘auto’ : Determine categories automatically from the training data.
list :
categories[i]holds the categories expected in the ith column. The passed categories should not mix strings and numeric values, and should be sorted in case of numeric values.
The used categories can be found in the
categories_attribute.- dtypenumber type, default np.float64
Desired dtype of output.
- handle_unknown{‘error’, ‘use_encoded_value’}, default=’error’
When set to ‘error’ an error will be raised in case an unknown categorical feature is present during transform. When set to ‘use_encoded_value’, the encoded value of unknown categories will be set to the value given for the parameter
unknown_value. Ininverse_transform, an unknown category will be denoted as None.New in version 0.24.
- unknown_valueint or np.nan, default=None
When the parameter handle_unknown is set to ‘use_encoded_value’, this parameter is required and will set the encoded value of unknown categories. It has to be distinct from the values used to encode any of the categories in
fit. If set to np.nan, thedtypeparameter must be a float dtype.New in version 0.24.
- encoded_missing_valueint or np.nan, default=np.nan
Encoded value of missing categories. If set to
np.nan, then thedtypeparameter must be a float dtype.New in version 1.1.
- Attributes:
- categories_list of arrays
The categories of each feature determined during
fit(in order of the features in X and corresponding with the output oftransform). This does not include categories that weren’t seen duringfit.- n_features_in_int
Number of features seen during fit.
New in version 1.0.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
OneHotEncoderPerforms a one-hot encoding of categorical features.
LabelEncoderEncodes target labels with values between 0 and
n_classes-1.
Notes
With a high proportion of
nanvalues, inferring categories becomes slow with Python versions before 3.10. The handling ofnanvalues was improved from Python 3.10 onwards, (c.f. bpo-43475).Examples
Given a dataset with two features, we let the encoder find the unique values per feature and transform the data to an ordinal encoding.
>>> from sklearn.preprocessing import OrdinalEncoder >>> enc = OrdinalEncoder() >>> X = [['Male', 1], ['Female', 3], ['Female', 2]] >>> enc.fit(X) OrdinalEncoder() >>> enc.categories_ [array(['Female', 'Male'], dtype=object), array([1, 2, 3], dtype=object)] >>> enc.transform([['Female', 3], ['Male', 1]]) array([[0., 2.], [1., 0.]])
>>> enc.inverse_transform([[1, 0], [0, 1]]) array([['Male', 1], ['Female', 2]], dtype=object)
By default,
OrdinalEncoderis lenient towards missing values by propagating them.>>> import numpy as np >>> X = [['Male', 1], ['Female', 3], ['Female', np.nan]] >>> enc.fit_transform(X) array([[ 1., 0.], [ 0., 1.], [ 0., nan]])
You can use the parameter
encoded_missing_valueto encode missing values.>>> enc.set_params(encoded_missing_value=-1).fit_transform(X) array([[ 1., 0.], [ 0., 1.], [ 0., -1.]])
Methods
fit(X[, y])Fit the OrdinalEncoder to X.
fit_transform(X[, y])Fit to data, then transform it.
get_feature_names_out([input_features])Get output feature names for transformation.
get_params([deep])Get parameters for this estimator.
Convert the data back to the original representation.
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform X to ordinal codes.
- fit(X, y=None)[source]¶
Fit the OrdinalEncoder to X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
The data to determine the categories of each feature.
- yNone
Ignored. This parameter exists only for compatibility with
Pipeline.
- Returns:
- selfobject
Fitted encoder.
- fit_transform(X, y=None, **fit_params)[source]¶
Fit to data, then transform it.
Fits transformer to
Xandywith optional parametersfit_paramsand returns a transformed version ofX.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs), default=None
Target values (None for unsupervised transformations).
- **fit_paramsdict
Additional fit parameters.
- Returns:
- X_newndarray array of shape (n_samples, n_features_new)
Transformed array.
- get_feature_names_out(input_features=None)[source]¶
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Same as input features.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- inverse_transform(X)[source]¶
Convert the data back to the original representation.
- Parameters:
- Xarray-like of shape (n_samples, n_encoded_features)
The transformed data.
- Returns:
- X_trndarray of shape (n_samples, n_features)
Inverse transformed array.
- set_output(*, transform=None)[source]¶
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame outputNone: Transform configuration is unchanged
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using sklearn.preprocessing.OrdinalEncoder¶
Partial Dependence and Individual Conditional Expectation Plots
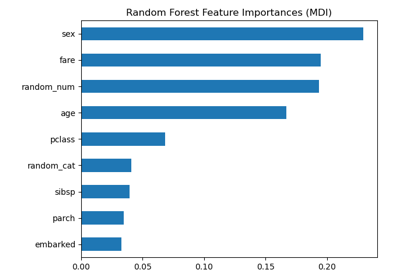
Permutation Importance vs Random Forest Feature Importance (MDI)