sklearn.model_selection.HalvingRandomSearchCV¶
- class sklearn.model_selection.HalvingRandomSearchCV(estimator, param_distributions, *, n_candidates='exhaust', factor=3, resource='n_samples', max_resources='auto', min_resources='smallest', aggressive_elimination=False, cv=5, scoring=None, refit=True, error_score=nan, return_train_score=True, random_state=None, n_jobs=None, verbose=0)[source]¶
Randomized search on hyper parameters.
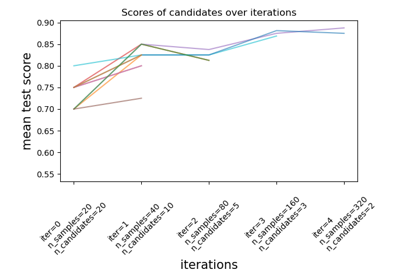
The search strategy starts evaluating all the candidates with a small amount of resources and iteratively selects the best candidates, using more and more resources.
The candidates are sampled at random from the parameter space and the number of sampled candidates is determined by
n_candidates.Read more in the User guide.
Note
This estimator is still experimental for now: the predictions and the API might change without any deprecation cycle. To use it, you need to explicitly import
enable_halving_search_cv:>>> # explicitly require this experimental feature >>> from sklearn.experimental import enable_halving_search_cv # noqa >>> # now you can import normally from model_selection >>> from sklearn.model_selection import HalvingRandomSearchCV
- Parameters
- estimatorestimator object
This is assumed to implement the scikit-learn estimator interface. Either estimator needs to provide a
scorefunction, orscoringmust be passed.- param_distributionsdict
Dictionary with parameters names (string) as keys and distributions or lists of parameters to try. Distributions must provide a
rvsmethod for sampling (such as those from scipy.stats.distributions). If a list is given, it is sampled uniformly.- n_candidatesint, default=’exhaust’
The number of candidate parameters to sample, at the first iteration. Using ‘exhaust’ will sample enough candidates so that the last iteration uses as many resources as possible, based on
min_resources,max_resourcesandfactor. In this case,min_resourcescannot be ‘exhaust’.- factorint or float, default=3
The ‘halving’ parameter, which determines the proportion of candidates that are selected for each subsequent iteration. For example,
factor=3means that only one third of the candidates are selected.- resource
'n_samples'or str, default=’n_samples’ Defines the resource that increases with each iteration. By default, the resource is the number of samples. It can also be set to any parameter of the base estimator that accepts positive integer values, e.g. ‘n_iterations’ or ‘n_estimators’ for a gradient boosting estimator. In this case
max_resourcescannot be ‘auto’ and must be set explicitly.- max_resourcesint, default=’auto’
The maximum number of resources that any candidate is allowed to use for a given iteration. By default, this is set
n_sampleswhenresource='n_samples'(default), else an error is raised.- min_resources{‘exhaust’, ‘smallest’} or int, default=’smallest’
The minimum amount of resource that any candidate is allowed to use for a given iteration. Equivalently, this defines the amount of resources
r0that are allocated for each candidate at the first iteration.‘smallest’ is a heuristic that sets
r0to a small value:n_splits * 2whenresource='n_samples'for a regression problemn_classes * n_splits * 2whenresource='n_samples'for a classification problem1whenresource != 'n_samples'
‘exhaust’ will set
r0such that the last iteration uses as much resources as possible. Namely, the last iteration will use the highest value smaller thanmax_resourcesthat is a multiple of bothmin_resourcesandfactor. In general, using ‘exhaust’ leads to a more accurate estimator, but is slightly more time consuming. ‘exhaust’ isn’t available whenn_candidates='exhaust'.
Note that the amount of resources used at each iteration is always a multiple of
min_resources.- aggressive_eliminationbool, default=False
This is only relevant in cases where there isn’t enough resources to reduce the remaining candidates to at most
factorafter the last iteration. IfTrue, then the search process will ‘replay’ the first iteration for as long as needed until the number of candidates is small enough. This isFalseby default, which means that the last iteration may evaluate more thanfactorcandidates. See Aggressive elimination of candidates for more details.- cvint, cross-validation generator or an iterable, default=5
Determines the cross-validation splitting strategy. Possible inputs for cv are:
integer, to specify the number of folds in a
(Stratified)KFold,An iterable yielding (train, test) splits as arrays of indices.
For integer/None inputs, if the estimator is a classifier and
yis either binary or multiclass,StratifiedKFoldis used. In all other cases,KFoldis used. These splitters are instantiated withshuffle=Falseso the splits will be the same across calls.Refer User Guide for the various cross-validation strategies that can be used here.
Note
Due to implementation details, the folds produced by
cvmust be the same across multiple calls tocv.split(). For built-inscikit-learniterators, this can be achieved by deactivating shuffling (shuffle=False), or by setting thecv’srandom_stateparameter to an integer.- scoringstr, callable, or None, default=None
A single string (see The scoring parameter: defining model evaluation rules) or a callable (see Defining your scoring strategy from metric functions) to evaluate the predictions on the test set. If None, the estimator’s score method is used.
- refitbool, default=True
If True, refit an estimator using the best found parameters on the whole dataset.
The refitted estimator is made available at the
best_estimator_attribute and permits usingpredictdirectly on thisHalvingRandomSearchCVinstance.- error_score‘raise’ or numeric
Value to assign to the score if an error occurs in estimator fitting. If set to ‘raise’, the error is raised. If a numeric value is given, FitFailedWarning is raised. This parameter does not affect the refit step, which will always raise the error. Default is
np.nan.- return_train_scorebool, default=False
If
False, thecv_results_attribute will not include training scores. Computing training scores is used to get insights on how different parameter settings impact the overfitting/underfitting trade-off. However computing the scores on the training set can be computationally expensive and is not strictly required to select the parameters that yield the best generalization performance.- random_stateint, RandomState instance or None, default=None
Pseudo random number generator state used for subsampling the dataset when
resources != 'n_samples'. Also used for random uniform sampling from lists of possible values instead of scipy.stats distributions. Pass an int for reproducible output across multiple function calls. See Glossary.- n_jobsint or None, default=None
Number of jobs to run in parallel.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- verboseint
Controls the verbosity: the higher, the more messages.
- Attributes
- n_resources_list of int
The amount of resources used at each iteration.
- n_candidates_list of int
The number of candidate parameters that were evaluated at each iteration.
- n_remaining_candidates_int
The number of candidate parameters that are left after the last iteration. It corresponds to
ceil(n_candidates[-1] / factor)- max_resources_int
The maximum number of resources that any candidate is allowed to use for a given iteration. Note that since the number of resources used at each iteration must be a multiple of
min_resources_, the actual number of resources used at the last iteration may be smaller thanmax_resources_.- min_resources_int
The amount of resources that are allocated for each candidate at the first iteration.
- n_iterations_int
The actual number of iterations that were run. This is equal to
n_required_iterations_ifaggressive_eliminationisTrue. Else, this is equal tomin(n_possible_iterations_, n_required_iterations_).- n_possible_iterations_int
The number of iterations that are possible starting with
min_resources_resources and without exceedingmax_resources_.- n_required_iterations_int
The number of iterations that are required to end up with less than
factorcandidates at the last iteration, starting withmin_resources_resources. This will be smaller thann_possible_iterations_when there isn’t enough resources.- cv_results_dict of numpy (masked) ndarrays
A dict with keys as column headers and values as columns, that can be imported into a pandas
DataFrame. It contains lots of information for analysing the results of a search. Please refer to the User guide for details.- best_estimator_estimator or dict
Estimator that was chosen by the search, i.e. estimator which gave highest score (or smallest loss if specified) on the left out data. Not available if
refit=False.- best_score_float
Mean cross-validated score of the best_estimator.
- best_params_dict
Parameter setting that gave the best results on the hold out data.
- best_index_int
The index (of the
cv_results_arrays) which corresponds to the best candidate parameter setting.The dict at
search.cv_results_['params'][search.best_index_]gives the parameter setting for the best model, that gives the highest mean score (search.best_score_).- scorer_function or a dict
Scorer function used on the held out data to choose the best parameters for the model.
- n_splits_int
The number of cross-validation splits (folds/iterations).
- refit_time_float
Seconds used for refitting the best model on the whole dataset.
This is present only if
refitis not False.- multimetric_bool
Whether or not the scorers compute several metrics.
classes_ndarray of shape (n_classes,)Class labels.
n_features_in_intNumber of features seen during fit.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Only defined if
best_estimator_is defined (see the documentation for therefitparameter for more details) and thatbest_estimator_exposesfeature_names_in_when fit.New in version 1.0.
See also
HalvingGridSearchCVSearch over a grid of parameters using successive halving.
Notes
The parameters selected are those that maximize the score of the held-out data, according to the scoring parameter.
Examples
>>> from sklearn.datasets import load_iris >>> from sklearn.ensemble import RandomForestClassifier >>> from sklearn.experimental import enable_halving_search_cv # noqa >>> from sklearn.model_selection import HalvingRandomSearchCV >>> from scipy.stats import randint >>> import numpy as np ... >>> X, y = load_iris(return_X_y=True) >>> clf = RandomForestClassifier(random_state=0) >>> np.random.seed(0) ... >>> param_distributions = {"max_depth": [3, None], ... "min_samples_split": randint(2, 11)} >>> search = HalvingRandomSearchCV(clf, param_distributions, ... resource='n_estimators', ... max_resources=10, ... random_state=0).fit(X, y) >>> search.best_params_ {'max_depth': None, 'min_samples_split': 10, 'n_estimators': 9}
Methods
Call decision_function on the estimator with the best found parameters.
fit(X[, y, groups])Run fit with all sets of parameters.
get_params([deep])Get parameters for this estimator.
Call inverse_transform on the estimator with the best found params.
predict(X)Call predict on the estimator with the best found parameters.
Call predict_log_proba on the estimator with the best found parameters.
Call predict_proba on the estimator with the best found parameters.
score(X[, y])Return the score on the given data, if the estimator has been refit.
Call score_samples on the estimator with the best found parameters.
set_params(**params)Set the parameters of this estimator.
transform(X)Call transform on the estimator with the best found parameters.
- property classes_¶
Class labels.
Only available when
refit=Trueand the estimator is a classifier.
- decision_function(X)[source]¶
Call decision_function on the estimator with the best found parameters.
Only available if
refit=Trueand the underlying estimator supportsdecision_function.- Parameters
- Xindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- y_scorendarray of shape (n_samples,) or (n_samples, n_classes) or (n_samples, n_classes * (n_classes-1) / 2)
Result of the decision function for
Xbased on the estimator with the best found parameters.
- fit(X, y=None, groups=None, **fit_params)[source]¶
Run fit with all sets of parameters.
- Parameters
- Xarray-like, shape (n_samples, n_features)
Training vector, where
n_samplesis the number of samples andn_featuresis the number of features.- yarray-like, shape (n_samples,) or (n_samples, n_output), optional
Target relative to X for classification or regression; None for unsupervised learning.
- groupsarray-like of shape (n_samples,), default=None
Group labels for the samples used while splitting the dataset into train/test set. Only used in conjunction with a “Group” cv instance (e.g.,
GroupKFold).- **fit_paramsdict of string -> object
Parameters passed to the
fitmethod of the estimator.
- Returns
- selfobject
Instance of fitted estimator.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsdict
Parameter names mapped to their values.
- inverse_transform(Xt)[source]¶
Call inverse_transform on the estimator with the best found params.
Only available if the underlying estimator implements
inverse_transformandrefit=True.- Parameters
- Xtindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- X{ndarray, sparse matrix} of shape (n_samples, n_features)
Result of the
inverse_transformfunction forXtbased on the estimator with the best found parameters.
- predict(X)[source]¶
Call predict on the estimator with the best found parameters.
Only available if
refit=Trueand the underlying estimator supportspredict.- Parameters
- Xindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- y_predndarray of shape (n_samples,)
The predicted labels or values for
Xbased on the estimator with the best found parameters.
- predict_log_proba(X)[source]¶
Call predict_log_proba on the estimator with the best found parameters.
Only available if
refit=Trueand the underlying estimator supportspredict_log_proba.- Parameters
- Xindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- y_predndarray of shape (n_samples,) or (n_samples, n_classes)
Predicted class log-probabilities for
Xbased on the estimator with the best found parameters. The order of the classes corresponds to that in the fitted attribute classes_.
- predict_proba(X)[source]¶
Call predict_proba on the estimator with the best found parameters.
Only available if
refit=Trueand the underlying estimator supportspredict_proba.- Parameters
- Xindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- y_predndarray of shape (n_samples,) or (n_samples, n_classes)
Predicted class probabilities for
Xbased on the estimator with the best found parameters. The order of the classes corresponds to that in the fitted attribute classes_.
- score(X, y=None)[source]¶
Return the score on the given data, if the estimator has been refit.
This uses the score defined by
scoringwhere provided, and thebest_estimator_.scoremethod otherwise.- Parameters
- Xarray-like of shape (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.- yarray-like of shape (n_samples, n_output) or (n_samples,), default=None
Target relative to X for classification or regression; None for unsupervised learning.
- Returns
- scorefloat
The score defined by
scoringif provided, and thebest_estimator_.scoremethod otherwise.
- score_samples(X)[source]¶
Call score_samples on the estimator with the best found parameters.
Only available if
refit=Trueand the underlying estimator supportsscore_samples.New in version 0.24.
- Parameters
- Xiterable
Data to predict on. Must fulfill input requirements of the underlying estimator.
- Returns
- y_scorendarray of shape (n_samples,)
The
best_estimator_.score_samplesmethod.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfestimator instance
Estimator instance.
- transform(X)[source]¶
Call transform on the estimator with the best found parameters.
Only available if the underlying estimator supports
transformandrefit=True.- Parameters
- Xindexable, length n_samples
Must fulfill the input assumptions of the underlying estimator.
- Returns
- Xt{ndarray, sparse matrix} of shape (n_samples, n_features)
Xtransformed in the new space based on the estimator with the best found parameters.