Note
Click here to download the full example code or to run this example in your browser via Binder
Scalable learning with polynomial kernel approximation¶
This example illustrates the use of PolynomialCountSketch to
efficiently generate polynomial kernel feature-space approximations.
This is used to train linear classifiers that approximate the accuracy
of kernelized ones.
We use the Covtype dataset [2], trying to reproduce the experiments on the
original paper of Tensor Sketch [1], i.e. the algorithm implemented by
PolynomialCountSketch.
First, we compute the accuracy of a linear classifier on the original
features. Then, we train linear classifiers on different numbers of
features (n_components) generated by PolynomialCountSketch,
approximating the accuracy of a kernelized classifier in a scalable manner.
# Author: Daniel Lopez-Sanchez <lope@usal.es>
# License: BSD 3 clause
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_covtype
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, Normalizer
from sklearn.svm import LinearSVC
from sklearn.kernel_approximation import PolynomialCountSketch
from sklearn.pipeline import Pipeline, make_pipeline
import time
Load the Covtype dataset, which contains 581,012 samples with 54 features each, distributed among 6 classes. The goal of this dataset is to predict forest cover type from cartographic variables only (no remotely sensed data). After loading, we transform it into a binary classification problem to match the version of the dataset in the LIBSVM webpage [2], which was the one used in [1].
X, y = fetch_covtype(return_X_y=True)
y[y != 2] = 0
y[y == 2] = 1 # We will try to separate class 2 from the other 6 classes.
Here we select 5,000 samples for training and 10,000 for testing. To actually reproduce the results in the original Tensor Sketch paper, select 100,000 for training.
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=5_000, test_size=10_000, random_state=42
)
Now scale features to the range [0, 1] to match the format of the dataset in the LIBSVM webpage, and then normalize to unit length as done in the original Tensor Sketch paper [1].
mm = make_pipeline(MinMaxScaler(), Normalizer())
X_train = mm.fit_transform(X_train)
X_test = mm.transform(X_test)
As a baseline, train a linear SVM on the original features and print the accuracy. We also measure and store accuracies and training times to plot them latter.
Out:
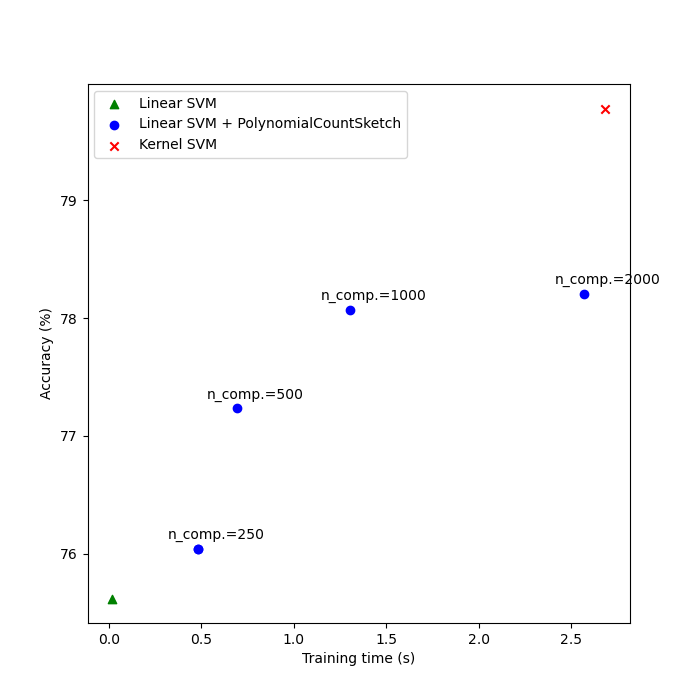
Linear SVM score on raw features: 75.62%
Then we train linear SVMs on the features generated by
PolynomialCountSketch with different values for n_components,
showing that these kernel feature approximations improve the accuracy
of linear classification. In typical application scenarios, n_components
should be larger than the number of features in the input representation
in order to achieve an improvement with respect to linear classification.
As a rule of thumb, the optimum of evaluation score / run time cost is
typically achieved at around n_components = 10 * n_features, though this
might depend on the specific dataset being handled. Note that, since the
original samples have 54 features, the explicit feature map of the
polynomial kernel of degree four would have approximately 8.5 million
features (precisely, 54^4). Thanks to PolynomialCountSketch, we can
condense most of the discriminative information of that feature space into a
much more compact representation. We repeat the experiment 5 times to
compensate for the stochastic nature of PolynomialCountSketch.
n_runs = 3
for n_components in [250, 500, 1000, 2000]:
ps_lsvm_time = 0
ps_lsvm_score = 0
for _ in range(n_runs):
pipeline = Pipeline(
steps=[
(
"kernel_approximator",
PolynomialCountSketch(n_components=n_components, degree=4),
),
("linear_classifier", LinearSVC()),
]
)
start = time.time()
pipeline.fit(X_train, y_train)
ps_lsvm_time += time.time() - start
ps_lsvm_score += 100 * pipeline.score(X_test, y_test)
ps_lsvm_time /= n_runs
ps_lsvm_score /= n_runs
results[f"LSVM + PS({n_components})"] = {
"time": ps_lsvm_time,
"score": ps_lsvm_score,
}
print(
f"Linear SVM score on {n_components} PolynomialCountSketch "
+ f"features: {ps_lsvm_score:.2f}%"
)
Out:
Linear SVM score on 250 PolynomialCountSketch features: 76.04%
Linear SVM score on 500 PolynomialCountSketch features: 77.23%
Linear SVM score on 1000 PolynomialCountSketch features: 78.07%
Linear SVM score on 2000 PolynomialCountSketch features: 78.20%
Train a kernelized SVM to see how well PolynomialCountSketch
is approximating the performance of the kernel. This, of course, may take
some time, as the SVC class has a relatively poor scalability. This is the
reason why kernel approximators are so useful:
from sklearn.svm import SVC
ksvm = SVC(C=500.0, kernel="poly", degree=4, coef0=0, gamma=1.0)
start = time.time()
ksvm.fit(X_train, y_train)
ksvm_time = time.time() - start
ksvm_score = 100 * ksvm.score(X_test, y_test)
results["KSVM"] = {"time": ksvm_time, "score": ksvm_score}
print(f"Kernel-SVM score on raw featrues: {ksvm_score:.2f}%")
Out:
Kernel-SVM score on raw featrues: 79.78%
Finally, plot the results of the different methods against their training times. As we can see, the kernelized SVM achieves a higher accuracy, but its training time is much larger and, most importantly, will grow much faster if the number of training samples increases.
N_COMPONENTS = [250, 500, 1000, 2000]
fig, ax = plt.subplots(figsize=(7, 7))
ax.scatter(
[
results["LSVM"]["time"],
],
[
results["LSVM"]["score"],
],
label="Linear SVM",
c="green",
marker="^",
)
ax.scatter(
[
results["LSVM + PS(250)"]["time"],
],
[
results["LSVM + PS(250)"]["score"],
],
label="Linear SVM + PolynomialCountSketch",
c="blue",
)
for n_components in N_COMPONENTS:
ax.scatter(
[
results[f"LSVM + PS({n_components})"]["time"],
],
[
results[f"LSVM + PS({n_components})"]["score"],
],
c="blue",
)
ax.annotate(
f"n_comp.={n_components}",
(
results[f"LSVM + PS({n_components})"]["time"],
results[f"LSVM + PS({n_components})"]["score"],
),
xytext=(-30, 10),
textcoords="offset pixels",
)
ax.scatter(
[
results["KSVM"]["time"],
],
[
results["KSVM"]["score"],
],
label="Kernel SVM",
c="red",
marker="x",
)
ax.set_xlabel("Training time (s)")
ax.set_ylabel("Accuracy (%)")
ax.legend()
plt.show()
References¶
[1] Pham, Ninh and Rasmus Pagh. “Fast and scalable polynomial kernels via explicit feature maps.” KDD ‘13 (2013). https://doi.org/10.1145/2487575.2487591
[2] LIBSVM binary datasets repository https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/binary.html
Total running time of the script: ( 0 minutes 33.319 seconds)
