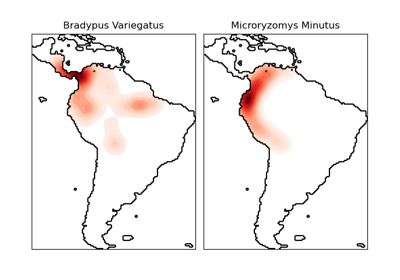
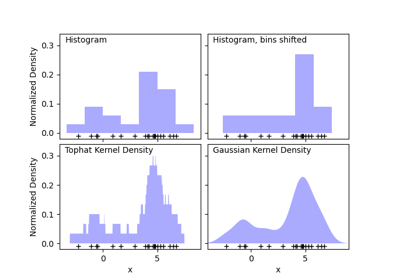
sklearn.neighbors.KernelDensity¶
-
class
sklearn.neighbors.KernelDensity(*, bandwidth=1.0, algorithm='auto', kernel='gaussian', metric='euclidean', atol=0, rtol=0, breadth_first=True, leaf_size=40, metric_params=None)[source]¶ Kernel Density Estimation.
Read more in the User Guide.
- Parameters
- bandwidthfloat, default=1.0
The bandwidth of the kernel.
- algorithm{‘kd_tree’, ‘ball_tree’, ‘auto’}, default=’auto’
The tree algorithm to use.
- kernel{‘gaussian’, ‘tophat’, ‘epanechnikov’, ‘exponential’, ‘linear’, ‘cosine’}, default=’gaussian’
The kernel to use.
- metricstr, default=’euclidean’
The distance metric to use. Note that not all metrics are valid with all algorithms. Refer to the documentation of
BallTreeandKDTreefor a description of available algorithms. Note that the normalization of the density output is correct only for the Euclidean distance metric. Default is ‘euclidean’.- atolfloat, default=0
The desired absolute tolerance of the result. A larger tolerance will generally lead to faster execution.
- rtolfloat, default=0
The desired relative tolerance of the result. A larger tolerance will generally lead to faster execution.
- breadth_firstbool, default=True
If true (default), use a breadth-first approach to the problem. Otherwise use a depth-first approach.
- leaf_sizeint, default=40
Specify the leaf size of the underlying tree. See
BallTreeorKDTreefor details.- metric_paramsdict, default=None
Additional parameters to be passed to the tree for use with the metric. For more information, see the documentation of
BallTreeorKDTree.
- Attributes
- tree_
BinaryTreeinstance The tree algorithm for fast generalized N-point problems.
- tree_
See also
sklearn.neighbors.KDTreeK-dimensional tree for fast generalized N-point problems.
sklearn.neighbors.BallTreeBall tree for fast generalized N-point problems.
Examples
Compute a gaussian kernel density estimate with a fixed bandwidth.
>>> import numpy as np >>> rng = np.random.RandomState(42) >>> X = rng.random_sample((100, 3)) >>> kde = KernelDensity(kernel='gaussian', bandwidth=0.5).fit(X) >>> log_density = kde.score_samples(X[:3]) >>> log_density array([-1.52955942, -1.51462041, -1.60244657])
Methods
fit(X[, y, sample_weight])Fit the Kernel Density model on the data.
get_params([deep])Get parameters for this estimator.
sample([n_samples, random_state])Generate random samples from the model.
score(X[, y])Compute the total log probability density under the model.
Evaluate the log density model on the data.
set_params(**params)Set the parameters of this estimator.
-
fit(X, y=None, sample_weight=None)[source]¶ Fit the Kernel Density model on the data.
- Parameters
- Xarray-like of shape (n_samples, n_features)
List of n_features-dimensional data points. Each row corresponds to a single data point.
- yNone
Ignored. This parameter exists only for compatibility with
Pipeline.- sample_weightarray-like of shape (n_samples,), default=None
List of sample weights attached to the data X.
New in version 0.20.
- Returns
- selfobject
Returns instance of object.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsdict
Parameter names mapped to their values.
-
sample(n_samples=1, random_state=None)[source]¶ Generate random samples from the model.
Currently, this is implemented only for gaussian and tophat kernels.
- Parameters
- n_samplesint, default=1
Number of samples to generate.
- random_stateint, RandomState instance or None, default=None
Determines random number generation used to generate random samples. Pass an int for reproducible results across multiple function calls. See :term:
Glossary <random_state>.
- Returns
- Xarray-like of shape (n_samples, n_features)
List of samples.
-
score(X, y=None)[source]¶ Compute the total log probability density under the model.
- Parameters
- Xarray-like of shape (n_samples, n_features)
List of n_features-dimensional data points. Each row corresponds to a single data point.
- yNone
Ignored. This parameter exists only for compatibility with
Pipeline.
- Returns
- logprobfloat
Total log-likelihood of the data in X. This is normalized to be a probability density, so the value will be low for high-dimensional data.
-
score_samples(X)[source]¶ Evaluate the log density model on the data.
- Parameters
- Xarray-like of shape (n_samples, n_features)
An array of points to query. Last dimension should match dimension of training data (n_features).
- Returns
- densityndarray of shape (n_samples,)
The array of log(density) evaluations. These are normalized to be probability densities, so values will be low for high-dimensional data.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfestimator instance
Estimator instance.