sklearn.decomposition.NMF¶
-
class
sklearn.decomposition.NMF(n_components=None, *, init='warn', solver='cd', beta_loss='frobenius', tol=0.0001, max_iter=200, random_state=None, alpha=0.0, l1_ratio=0.0, verbose=0, shuffle=False, regularization='both')[source]¶ Non-Negative Matrix Factorization (NMF).
Find two non-negative matrices (W, H) whose product approximates the non- negative matrix X. This factorization can be used for example for dimensionality reduction, source separation or topic extraction.
The objective function is:
\[ \begin{align}\begin{aligned}0.5 * ||X - WH||_{loss}^2 + alpha * l1_{ratio} * ||vec(W)||_1\\+ alpha * l1_{ratio} * ||vec(H)||_1\\+ 0.5 * alpha * (1 - l1_{ratio}) * ||W||_{Fro}^2\\+ 0.5 * alpha * (1 - l1_{ratio}) * ||H||_{Fro}^2\end{aligned}\end{align} \]Where:
\(||A||_{Fro}^2 = \sum_{i,j} A_{ij}^2\) (Frobenius norm)
\(||vec(A)||_1 = \sum_{i,j} abs(A_{ij})\) (Elementwise L1 norm)
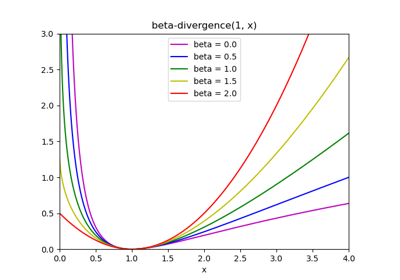
The generic norm \(||X - WH||_{loss}\) may represent the Frobenius norm or another supported beta-divergence loss. The choice between options is controlled by the
beta_lossparameter.The objective function is minimized with an alternating minimization of W and H.
Read more in the User Guide.
- Parameters
- n_componentsint, default=None
Number of components, if n_components is not set all features are kept.
- init{‘random’, ‘nndsvd’, ‘nndsvda’, ‘nndsvdar’, ‘custom’}, default=None
Method used to initialize the procedure. Default: None. Valid options:
None: ‘nndsvd’ if n_components <= min(n_samples, n_features), otherwise random.'random': non-negative random matrices, scaled with: sqrt(X.mean() / n_components)'nndsvd': Nonnegative Double Singular Value Decomposition (NNDSVD) initialization (better for sparseness)'nndsvda': NNDSVD with zeros filled with the average of X (better when sparsity is not desired)'nndsvdar'NNDSVD with zeros filled with small random values (generally faster, less accurate alternative to NNDSVDa for when sparsity is not desired)'custom': use custom matrices W and H
- solver{‘cd’, ‘mu’}, default=’cd’
Numerical solver to use: ‘cd’ is a Coordinate Descent solver. ‘mu’ is a Multiplicative Update solver.
New in version 0.17: Coordinate Descent solver.
New in version 0.19: Multiplicative Update solver.
- beta_lossfloat or {‘frobenius’, ‘kullback-leibler’, ‘itakura-saito’}, default=’frobenius’
Beta divergence to be minimized, measuring the distance between X and the dot product WH. Note that values different from ‘frobenius’ (or 2) and ‘kullback-leibler’ (or 1) lead to significantly slower fits. Note that for beta_loss <= 0 (or ‘itakura-saito’), the input matrix X cannot contain zeros. Used only in ‘mu’ solver.
New in version 0.19.
- tolfloat, default=1e-4
Tolerance of the stopping condition.
- max_iterint, default=200
Maximum number of iterations before timing out.
- random_stateint, RandomState instance or None, default=None
Used for initialisation (when
init== ‘nndsvdar’ or ‘random’), and in Coordinate Descent. Pass an int for reproducible results across multiple function calls. See Glossary.- alphafloat, default=0.
Constant that multiplies the regularization terms. Set it to zero to have no regularization.
New in version 0.17: alpha used in the Coordinate Descent solver.
- l1_ratiofloat, default=0.
The regularization mixing parameter, with 0 <= l1_ratio <= 1. For l1_ratio = 0 the penalty is an elementwise L2 penalty (aka Frobenius Norm). For l1_ratio = 1 it is an elementwise L1 penalty. For 0 < l1_ratio < 1, the penalty is a combination of L1 and L2.
New in version 0.17: Regularization parameter l1_ratio used in the Coordinate Descent solver.
- verboseint, default=0
Whether to be verbose.
- shufflebool, default=False
If true, randomize the order of coordinates in the CD solver.
New in version 0.17: shuffle parameter used in the Coordinate Descent solver.
- regularization{‘both’, ‘components’, ‘transformation’, None}, default=’both’
Select whether the regularization affects the components (H), the transformation (W), both or none of them.
New in version 0.24.
- Attributes
- components_ndarray of shape (n_components, n_features)
Factorization matrix, sometimes called ‘dictionary’.
- n_components_int
The number of components. It is same as the
n_componentsparameter if it was given. Otherwise, it will be same as the number of features.- reconstruction_err_float
Frobenius norm of the matrix difference, or beta-divergence, between the training data
Xand the reconstructed dataWHfrom the fitted model.- n_iter_int
Actual number of iterations.
References
Cichocki, Andrzej, and P. H. A. N. Anh-Huy. “Fast local algorithms for large scale nonnegative matrix and tensor factorizations.” IEICE transactions on fundamentals of electronics, communications and computer sciences 92.3: 708-721, 2009.
Fevotte, C., & Idier, J. (2011). Algorithms for nonnegative matrix factorization with the beta-divergence. Neural Computation, 23(9).
Examples
>>> import numpy as np >>> X = np.array([[1, 1], [2, 1], [3, 1.2], [4, 1], [5, 0.8], [6, 1]]) >>> from sklearn.decomposition import NMF >>> model = NMF(n_components=2, init='random', random_state=0) >>> W = model.fit_transform(X) >>> H = model.components_
Methods
fit(X[, y])Learn a NMF model for the data X.
fit_transform(X[, y, W, H])Learn a NMF model for the data X and returns the transformed data.
get_params([deep])Get parameters for this estimator.
Transform data back to its original space.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform the data X according to the fitted NMF model.
-
fit(X, y=None, **params)[source]¶ Learn a NMF model for the data X.
- Parameters
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Data matrix to be decomposed
- yIgnored
- Returns
- self
-
fit_transform(X, y=None, W=None, H=None)[source]¶ Learn a NMF model for the data X and returns the transformed data.
This is more efficient than calling fit followed by transform.
- Parameters
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Data matrix to be decomposed
- yIgnored
- Warray-like of shape (n_samples, n_components)
If init=’custom’, it is used as initial guess for the solution.
- Harray-like of shape (n_components, n_features)
If init=’custom’, it is used as initial guess for the solution.
- Returns
- Wndarray of shape (n_samples, n_components)
Transformed data.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsdict
Parameter names mapped to their values.
-
inverse_transform(W)[source]¶ Transform data back to its original space.
- Parameters
- W{ndarray, sparse matrix} of shape (n_samples, n_components)
Transformed data matrix.
- Returns
- X{ndarray, sparse matrix} of shape (n_samples, n_features)
Data matrix of original shape.
New in version 0.18: ..
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfestimator instance
Estimator instance.