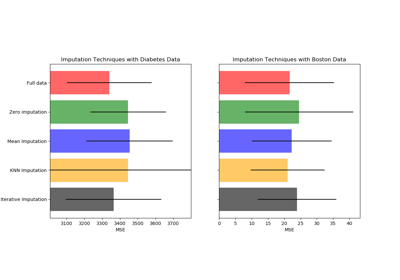
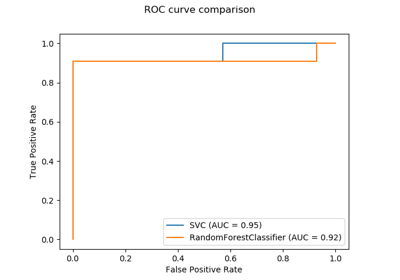
sklearn.impute.KNNImputer¶
-
class
sklearn.impute.KNNImputer(missing_values=nan, n_neighbors=5, weights='uniform', metric='nan_euclidean', copy=True, add_indicator=False)[source]¶ Imputation for completing missing values using k-Nearest Neighbors.
Each sample’s missing values are imputed using the mean value from
n_neighborsnearest neighbors found in the training set. Two samples are close if the features that neither is missing are close.Read more in the User Guide.
New in version 0.22.
- Parameters
- missing_valuesnumber, string, np.nan or None, default=`np.nan`
The placeholder for the missing values. All occurrences of
missing_valueswill be imputed.- n_neighborsint, default=5
Number of neighboring samples to use for imputation.
- weights{‘uniform’, ‘distance’} or callable, default=’uniform’
Weight function used in prediction. Possible values:
‘uniform’ : uniform weights. All points in each neighborhood are weighted equally.
‘distance’ : weight points by the inverse of their distance. in this case, closer neighbors of a query point will have a greater influence than neighbors which are further away.
callable : a user-defined function which accepts an array of distances, and returns an array of the same shape containing the weights.
- metric{‘nan_euclidean’} or callable, default=’nan_euclidean’
Distance metric for searching neighbors. Possible values:
‘nan_euclidean’
callable : a user-defined function which conforms to the definition of
_pairwise_callable(X, Y, metric, **kwds). The function accepts two arrays, X and Y, and amissing_valueskeyword inkwdsand returns a scalar distance value.
- copybool, default=True
If True, a copy of X will be created. If False, imputation will be done in-place whenever possible.
- add_indicatorbool, default=False
If True, a
MissingIndicatortransform will stack onto the output of the imputer’s transform. This allows a predictive estimator to account for missingness despite imputation. If a feature has no missing values at fit/train time, the feature won’t appear on the missing indicator even if there are missing values at transform/test time.
- Attributes
- indicator_
sklearn.impute.MissingIndicator Indicator used to add binary indicators for missing values.
Noneif add_indicator is False.
- indicator_
References
Olga Troyanskaya, Michael Cantor, Gavin Sherlock, Pat Brown, Trevor Hastie, Robert Tibshirani, David Botstein and Russ B. Altman, Missing value estimation methods for DNA microarrays, BIOINFORMATICS Vol. 17 no. 6, 2001 Pages 520-525.
Examples
>>> import numpy as np >>> from sklearn.impute import KNNImputer >>> X = [[1, 2, np.nan], [3, 4, 3], [np.nan, 6, 5], [8, 8, 7]] >>> imputer = KNNImputer(n_neighbors=2) >>> imputer.fit_transform(X) array([[1. , 2. , 4. ], [3. , 4. , 3. ], [5.5, 6. , 5. ], [8. , 8. , 7. ]])
Methods
fit(self, X[, y])Fit the imputer on X.
fit_transform(self, X[, y])Fit to data, then transform it.
get_params(self[, deep])Get parameters for this estimator.
set_params(self, \*\*params)Set the parameters of this estimator.
transform(self, X)Impute all missing values in X.
-
__init__(self, missing_values=nan, n_neighbors=5, weights='uniform', metric='nan_euclidean', copy=True, add_indicator=False)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(self, X, y=None)[source]¶ Fit the imputer on X.
- Parameters
- Xarray-like shape of (n_samples, n_features)
Input data, where
n_samplesis the number of samples andn_featuresis the number of features.
- Returns
- selfobject
-
fit_transform(self, X, y=None, **fit_params)[source]¶ Fit to data, then transform it.
Fits transformer to X and y with optional parameters fit_params and returns a transformed version of X.
- Parameters
- Xnumpy array of shape [n_samples, n_features]
Training set.
- ynumpy array of shape [n_samples]
Target values.
- **fit_paramsdict
Additional fit parameters.
- Returns
- X_newnumpy array of shape [n_samples, n_features_new]
Transformed array.
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsmapping of string to any
Parameter names mapped to their values.
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfobject
Estimator instance.
-
transform(self, X)[source]¶ Impute all missing values in X.
- Parameters
- Xarray-like of shape (n_samples, n_features)
The input data to complete.
- Returns
- Xarray-like of shape (n_samples, n_output_features)
The imputed dataset.
n_output_featuresis the number of features that is not always missing duringfit.