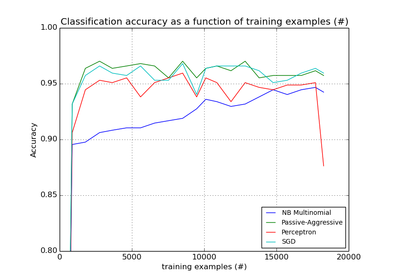
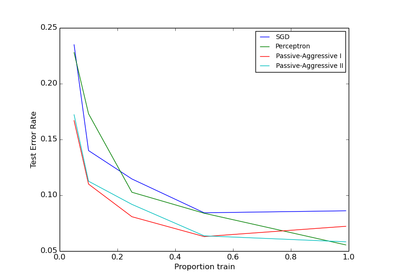
sklearn.linear_model.PassiveAggressiveClassifier¶
- class sklearn.linear_model.PassiveAggressiveClassifier(C=1.0, fit_intercept=True, n_iter=5, shuffle=False, verbose=0, loss='hinge', n_jobs=1, random_state=None, warm_start=False)¶
Passive Aggressive Classifier
Parameters: C : float
Maximum step size (regularization). Defaults to 1.0.
fit_intercept: bool :
Whether the intercept should be estimated or not. If False, the data is assumed to be already centered. Defaults to True.
n_iter: int, optional :
The number of passes over the training data (aka epochs). Defaults to 5.
shuffle: bool, optional :
Whether or not the training data should be shuffled after each epoch. Defaults to False.
random_state: int seed, RandomState instance, or None (default) :
The seed of the pseudo random number generator to use when shuffling the data.
verbose: integer, optional :
The verbosity level
n_jobs: integer, optional :
The number of CPUs to use to do the OVA (One Versus All, for multi-class problems) computation. -1 means ‘all CPUs’. Defaults to 1.
loss : string, optional
The loss function to be used: hinge: equivalent to PA-I in the reference paper. squared_hinge: equivalent to PA-II in the reference paper.
warm_start : bool, optional
When set to True, reuse the solution of the previous call to fit as initialization, otherwise, just erase the previous solution.
Attributes: `coef_` : array, shape = [1, n_features] if n_classes == 2 else [n_classes,
n_features] :
Weights assigned to the features.
`intercept_` : array, shape = [1] if n_classes == 2 else [n_classes]
Constants in decision function.
See also
References
Online Passive-Aggressive Algorithms <http://jmlr.csail.mit.edu/papers/volume7/crammer06a/crammer06a.pdf> K. Crammer, O. Dekel, J. Keshat, S. Shalev-Shwartz, Y. Singer - JMLR (2006)
Methods
decision_function(X) Predict confidence scores for samples. densify() Convert coefficient matrix to dense array format. fit(X, y[, coef_init, intercept_init]) Fit linear model with Passive Aggressive algorithm. get_params([deep]) Get parameters for this estimator. partial_fit(X, y[, classes]) Fit linear model with Passive Aggressive algorithm. predict(X) Predict class labels for samples in X. score(X, y[, sample_weight]) Returns the mean accuracy on the given test data and labels. set_params(*args, **kwargs) sparsify() Convert coefficient matrix to sparse format. - __init__(C=1.0, fit_intercept=True, n_iter=5, shuffle=False, verbose=0, loss='hinge', n_jobs=1, random_state=None, warm_start=False)¶
- decision_function(X)¶
Predict confidence scores for samples.
The confidence score for a sample is the signed distance of that sample to the hyperplane.
Parameters: X : {array-like, sparse matrix}, shape = (n_samples, n_features)
Samples.
Returns: array, shape=(n_samples,) if n_classes == 2 else (n_samples, n_classes) :
Confidence scores per (sample, class) combination. In the binary case, confidence score for self.classes_[1] where >0 means this class would be predicted.
- densify()¶
Convert coefficient matrix to dense array format.
Converts the coef_ member (back) to a numpy.ndarray. This is the default format of coef_ and is required for fitting, so calling this method is only required on models that have previously been sparsified; otherwise, it is a no-op.
Returns: self: estimator :
- fit(X, y, coef_init=None, intercept_init=None)¶
Fit linear model with Passive Aggressive algorithm.
Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
Training data
y : numpy array of shape [n_samples]
Target values
coef_init : array, shape = [n_classes,n_features]
The initial coefficients to warm-start the optimization.
intercept_init : array, shape = [n_classes]
The initial intercept to warm-start the optimization.
sample_weight : array-like, shape = [n_samples], optional
Weights applied to individual samples. If not provided, uniform weights are assumed.
Returns: self : returns an instance of self.
- get_params(deep=True)¶
Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
- partial_fit(X, y, classes=None)¶
Fit linear model with Passive Aggressive algorithm.
Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
Subset of the training data
y : numpy array of shape [n_samples]
Subset of the target values
classes : array, shape = [n_classes]
Classes across all calls to partial_fit. Can be obtained by via np.unique(y_all), where y_all is the target vector of the entire dataset. This argument is required for the first call to partial_fit and can be omitted in the subsequent calls. Note that y doesn’t need to contain all labels in classes.
Returns: self : returns an instance of self.
- predict(X)¶
Predict class labels for samples in X.
Parameters: X : {array-like, sparse matrix}, shape = [n_samples, n_features]
Samples.
Returns: C : array, shape = [n_samples]
Predicted class label per sample.
- score(X, y, sample_weight=None)¶
Returns the mean accuracy on the given test data and labels.
Parameters: X : array-like, shape = (n_samples, n_features)
Test samples.
y : array-like, shape = (n_samples,)
True labels for X.
sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns: score : float
Mean accuracy of self.predict(X) wrt. y.
- sparsify()¶
Convert coefficient matrix to sparse format.
Converts the coef_ member to a scipy.sparse matrix, which for L1-regularized models can be much more memory- and storage-efficient than the usual numpy.ndarray representation.
The intercept_ member is not converted.
Returns: self: estimator : Notes
For non-sparse models, i.e. when there are not many zeros in coef_, this may actually increase memory usage, so use this method with care. A rule of thumb is that the number of zero elements, which can be computed with (coef_ == 0).sum(), must be more than 50% for this to provide significant benefits.
After calling this method, further fitting with the partial_fit method (if any) will not work until you call densify.