Lasso#
- class sklearn.linear_model.Lasso(alpha=1.0, *, fit_intercept=True, precompute=False, copy_X=True, max_iter=1000, tol=0.0001, warm_start=False, positive=False, random_state=None, selection='cyclic')[source]#
Linear Model trained with L1 prior as regularizer (aka the Lasso).
The optimization objective for Lasso is:
(1 / (2 * n_samples)) * ||y - Xw||^2_2 + alpha * ||w||_1
Technically the Lasso model is optimizing the same objective function as the Elastic Net with
l1_ratio=1.0(no L2 penalty).Read more in the User Guide.
- Parameters:
- alphafloat, default=1.0
Constant that multiplies the L1 term, controlling regularization strength.
alphamust be a non-negative float i.e. in[0, inf).When
alpha = 0, the objective is equivalent to ordinary least squares, solved by theLinearRegressionobject. For numerical reasons, usingalpha = 0with theLassoobject is not advised. Instead, you should use theLinearRegressionobject.- fit_interceptbool, default=True
Whether to calculate the intercept for this model. If set to False, no intercept will be used in calculations (i.e. data is expected to be centered).
- precomputebool or array-like of shape (n_features, n_features), default=False
Whether to use a precomputed Gram matrix to speed up calculations. The Gram matrix can also be passed as argument. For sparse input this option is always
Falseto preserve sparsity.- copy_Xbool, default=True
If
True, X will be copied; else, it may be overwritten.- max_iterint, default=1000
The maximum number of iterations.
- tolfloat, default=1e-4
The tolerance for the optimization: if the updates are smaller or equal to
tol, the optimization code checks the dual gap for optimality and continues until it is smaller or equal totol, see Notes below.- warm_startbool, default=False
When set to
True, reuse the solution of the previous call to fit as initialization, otherwise, just erase the previous solution. See the Glossary.- positivebool, default=False
When set to
True, forces the coefficients to be positive.- random_stateint, RandomState instance, default=None
The seed of the pseudo random number generator that selects a random feature to update. Used when
selection== ‘random’. Pass an int for reproducible output across multiple function calls. See Glossary.- selection{‘cyclic’, ‘random’}, default=’cyclic’
If set to ‘random’, a random coefficient is updated every iteration rather than looping over features sequentially by default. This (setting to ‘random’) often leads to significantly faster convergence especially when tol is higher than 1e-4.
- Attributes:
- coef_ndarray of shape (n_features,) or (n_targets, n_features)
Parameter vector (w in the cost function formula).
- dual_gap_float or ndarray of shape (n_targets,)
Given param alpha, the dual gaps at the end of the optimization, same shape as each observation of y.
sparse_coef_sparse matrix of shape (n_features, 1) or (n_targets, n_features)Sparse representation of the fitted
coef_.- intercept_float or ndarray of shape (n_targets,)
Independent term in decision function.
- n_iter_int or list of int
Number of iterations run by the coordinate descent solver to reach the specified tolerance.
- n_features_in_int
Number of features seen during fit.
Added in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.Added in version 1.0.
See also
lars_pathRegularization path using LARS.
lasso_pathRegularization path using Lasso.
LassoLarsLasso Path along the regularization parameter using LARS algorithm.
LassoCVLasso alpha parameter by cross-validation.
LassoLarsCVLasso least angle parameter algorithm by cross-validation.
sklearn.decomposition.sparse_encodeSparse coding array estimator.
Notes
The algorithm used to fit the model is coordinate descent.
To avoid unnecessary memory duplication the X argument of the fit method should be directly passed as a Fortran-contiguous numpy array.
Regularization improves the conditioning of the problem and reduces the variance of the estimates. Larger values specify stronger regularization. Alpha corresponds to
1 / (2C)in other linear models such asLogisticRegressionorLinearSVC.The precise stopping criteria based on
tolare the following: First, check that that maximum coordinate update, i.e. \(\max_j |w_j^{new} - w_j^{old}|\) is smaller or equal totoltimes the maximum absolute coefficient, \(\max_j |w_j|\). If so, then additionally check whether the dual gap is smaller or equal totoltimes \(||y||_2^2 / n_{\text{samples}}\).The target can be a 2-dimensional array, resulting in the optimization of the following objective:
(1 / (2 * n_samples)) * ||Y - XW||^2_F + alpha * ||W||_11
where \(||W||_{1,1}\) is the sum of the magnitude of the matrix coefficients. It should not be confused with
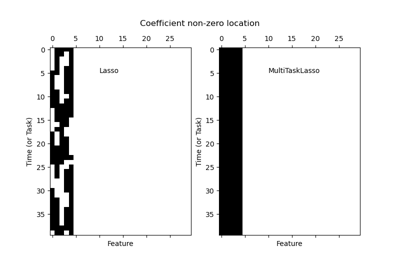
MultiTaskLassowhich instead penalizes the \(L_{2,1}\) norm of the coefficients, yielding row-wise sparsity in the coefficients.The underlying coordinate descent solver uses gap safe screening rules to speedup fitting time, see User Guide on coordinate descent.
Examples
>>> from sklearn import linear_model >>> clf = linear_model.Lasso(alpha=0.1) >>> clf.fit([[0,0], [1, 1], [2, 2]], [0, 1, 2]) Lasso(alpha=0.1) >>> print(clf.coef_) [0.85 0. ] >>> print(clf.intercept_) 0.15
L1-based models for Sparse Signals compares Lasso with other L1-based regression models (ElasticNet and ARD Regression) for sparse signal recovery in the presence of noise and feature correlation.
- fit(X, y, sample_weight=None, check_input=True)[source]#
Fit model with coordinate descent.
- Parameters:
- X{ndarray, sparse matrix, sparse array} of (n_samples, n_features)
Data.
Note that large sparse matrices and arrays requiring
int64indices are not accepted.- yndarray of shape (n_samples,) or (n_samples, n_targets)
Target. Will be cast to X’s dtype if necessary.
- sample_weightfloat or array-like of shape (n_samples,), default=None
Sample weights. Internally, the
sample_weightvector will be rescaled to sum ton_samples.Added in version 0.23.
- check_inputbool, default=True
Allow to bypass several input checking. Don’t use this parameter unless you know what you do.
- Returns:
- selfobject
Fitted estimator.
Notes
Coordinate descent is an algorithm that considers each column of data at a time hence it will automatically convert the X input as a Fortran-contiguous numpy array if necessary.
To avoid memory re-allocation it is advised to allocate the initial data in memory directly using that format.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)[source]#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- static path(X, y, *, l1_ratio=0.5, eps=0.001, n_alphas=100, alphas=None, precompute='auto', Xy=None, copy_X=True, coef_init=None, verbose=False, return_n_iter=False, positive=False, check_input=True, **params)[source]#
Compute elastic net path with coordinate descent.
The elastic net optimization function varies for mono and multi-outputs.
For mono-output tasks it is:
1 / (2 * n_samples) * ||y - Xw||^2_2 + alpha * l1_ratio * ||w||_1 + 0.5 * alpha * (1 - l1_ratio) * ||w||^2_2
For multi-output tasks it is:
(1 / (2 * n_samples)) * ||Y - XW||_Fro^2 + alpha * l1_ratio * ||W||_21 + 0.5 * alpha * (1 - l1_ratio) * ||W||_Fro^2
Where:
||W||_21 = \sum_i \sqrt{\sum_j w_{ij}^2}
i.e. the sum of norm of each row.
Read more in the User Guide.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Training data. Pass directly as Fortran-contiguous data to avoid unnecessary memory duplication. If
yis mono-output thenXcan be sparse.- y{array-like, sparse matrix} of shape (n_samples,) or (n_samples, n_targets)
Target values.
- l1_ratiofloat, default=0.5
Number between 0 and 1 passed to elastic net (scaling between l1 and l2 penalties).
l1_ratio=1corresponds to the Lasso.- epsfloat, default=1e-3
Length of the path.
eps=1e-3means thatalpha_min / alpha_max = 1e-3.- n_alphasint, default=100
Number of alphas along the regularization path.
- alphasarray-like, default=None
List of alphas where to compute the models. If None alphas are set automatically.
- precompute‘auto’, bool or array-like of shape (n_features, n_features), default=’auto’
Whether to use a precomputed Gram matrix to speed up calculations. If set to
'auto'let us decide. The Gram matrix can also be passed as argument.- Xyarray-like of shape (n_features,) or (n_features, n_targets), default=None
Xy = np.dot(X.T, y) that can be precomputed. It is useful only when the Gram matrix is precomputed.
- copy_Xbool, default=True
If
True, X will be copied; else, it may be overwritten.- coef_initarray-like of shape (n_features, ), default=None
The initial values of the coefficients.
- verbosebool or int, default=False
Amount of verbosity.
- return_n_iterbool, default=False
Whether to return the number of iterations or not.
- positivebool, default=False
If set to True, forces coefficients to be positive. (Only allowed when
y.ndim == 1).- check_inputbool, default=True
If set to False, the input validation checks are skipped (including the Gram matrix when provided). It is assumed that they are handled by the caller.
- **paramskwargs
Keyword arguments passed to the coordinate descent solver.
- Returns:
- alphasndarray of shape (n_alphas,)
The alphas along the path where models are computed.
- coefsndarray of shape (n_features, n_alphas) or (n_targets, n_features, n_alphas)
Coefficients along the path.
- dual_gapsndarray of shape (n_alphas,)
The dual gaps at the end of the optimization for each alpha.
- n_iterslist of int
The number of iterations taken by the coordinate descent optimizer to reach the specified tolerance for each alpha. (Is returned when
return_n_iteris set to True).
See also
MultiTaskElasticNetMulti-task ElasticNet model trained with L1/L2 mixed-norm as regularizer.
MultiTaskElasticNetCVMulti-task L1/L2 ElasticNet with built-in cross-validation.
ElasticNetLinear regression with combined L1 and L2 priors as regularizer.
ElasticNetCVElastic Net model with iterative fitting along a regularization path.
Notes
For an example, see examples/linear_model/plot_lasso_lasso_lars_elasticnet_path.py.
The underlying coordinate descent solver uses gap safe screening rules to speedup fitting time, see User Guide on coordinate descent.
Examples
>>> from sklearn.linear_model import enet_path >>> from sklearn.datasets import make_regression >>> X, y, true_coef = make_regression( ... n_samples=100, n_features=5, n_informative=2, coef=True, random_state=0 ... ) >>> true_coef array([ 0. , 0. , 0. , 97.9, 45.7]) >>> alphas, estimated_coef, _ = enet_path(X, y, n_alphas=3) >>> alphas.shape (3,) >>> estimated_coef array([[ 0., 0.787, 0.568], [ 0., 1.120, 0.620], [-0., -2.129, -1.128], [ 0., 23.046, 88.939], [ 0., 10.637, 41.566]])
- predict(X)[source]#
Predict using the linear model.
- Parameters:
- Xarray-like or sparse matrix, shape (n_samples, n_features)
Samples.
- Returns:
- Carray, shape (n_samples,)
Returns predicted values.
- score(X, y, sample_weight=None)[source]#
Return coefficient of determination on test data.
The coefficient of determination, \(R^2\), is defined as \((1 - \frac{u}{v})\), where \(u\) is the residual sum of squares
((y_true - y_pred)** 2).sum()and \(v\) is the total sum of squares((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value ofy, disregarding the input features, would get a \(R^2\) score of 0.0.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Test samples. For some estimators this may be a precomputed kernel matrix or a list of generic objects instead with shape
(n_samples, n_samples_fitted), wheren_samples_fittedis the number of samples used in the fitting for the estimator.- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True values for
X.- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns:
- scorefloat
\(R^2\) of
self.predict(X)w.r.t.y.
Notes
The \(R^2\) score used when calling
scoreon a regressor usesmultioutput='uniform_average'from version 0.23 to keep consistent with default value ofr2_score. This influences thescoremethod of all the multioutput regressors (except forMultiOutputRegressor).
- set_fit_request(*, sample_weight: bool | None | str = '$UNCHANGED$') Lasso[source]#
Configure whether metadata should be requested to be passed to the
fitmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- set_score_request(*, sample_weight: bool | None | str = '$UNCHANGED$') Lasso[source]#
Configure whether metadata should be requested to be passed to the
scoremethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toscoreif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toscore.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter inscore.
- Returns:
- selfobject
The updated object.
Gallery examples#
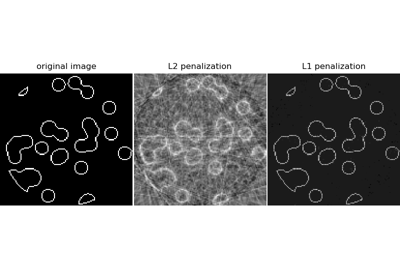
Compressive sensing: tomography reconstruction with L1 prior (Lasso)