Installing the development version of scikit-learn¶
This section introduces how to install the main branch of scikit-learn. This can be done by either installing a nightly build or building from source.
Installing nightly builds¶
The continuous integration servers of the scikit-learn project build, test and upload wheel packages for the most recent Python version on a nightly basis.
Installing a nightly build is the quickest way to:
try a new feature that will be shipped in the next release (that is, a feature from a pull-request that was recently merged to the main branch);
check whether a bug you encountered has been fixed since the last release.
You can install the nightly build of scikit-learn using the scientific-python-nightly-wheels
index from the PyPI registry of anaconda.org:
pip install --pre --extra-index https://pypi.anaconda.org/scientific-python-nightly-wheels/simple scikit-learn
Note that first uninstalling scikit-learn might be required to be able to install nightly builds of scikit-learn.
Building from source¶
Building from source is required to work on a contribution (bug fix, new feature, code or documentation improvement).
Use Git to check out the latest source from the scikit-learn repository on Github.:
git clone git://github.com/scikit-learn/scikit-learn.git # add --depth 1 if your connection is slow cd scikit-learn
If you plan on submitting a pull-request, you should clone from your fork instead.
Install a recent version of Python (3.9 is recommended at the time of writing) for instance using Miniforge3. Miniforge provides a conda-based distribution of Python and the most popular scientific libraries.
If you installed Python with conda, we recommend to create a dedicated conda environment with all the build dependencies of scikit-learn (namely NumPy, SciPy, and Cython):
conda create -n sklearn-env -c conda-forge python=3.9 numpy scipy cythonIt is not always necessary but it is safer to open a new prompt before activating the newly created conda environment.
conda activate sklearn-envAlternative to conda: If you run Linux or similar, you can instead use your system’s Python provided it is recent enough (3.8 or higher at the time of writing). In this case, we recommend to create a dedicated virtualenv and install the scikit-learn build dependencies with pip:
python3 -m venv sklearn-env source sklearn-env/bin/activate pip install wheel numpy scipy cython
Install a compiler with OpenMP support for your platform. See instructions for Windows, macOS, Linux and FreeBSD.
Build the project with pip in Editable mode:
pip install -v --no-use-pep517 --no-build-isolation -e .Check that the installed scikit-learn has a version number ending with
.dev0:python -c "import sklearn; sklearn.show_versions()"Please refer to the Developer’s Guide and Useful pytest aliases and flags to run the tests on the module of your choice.
Note
You will have to run the pip install -v --no-use-pep517 --no-build-isolation -e .
command every time the source code of a Cython file is updated
(ending in .pyx or .pxd). This can happen when you edit them or when you
use certain git commands such as git pull. Use the --no-build-isolation flag
to avoid compiling the whole project each time, only the files you have
modified. Include the --no-use-pep517 flag because the --no-build-isolation
option might not work otherwise (this is due to a bug which will be fixed in the
future).
Dependencies¶
Runtime dependencies¶
Scikit-learn requires the following dependencies both at build time and at runtime:
Python (>= 3.8),
NumPy (>= 1.17.3),
SciPy (>= 1.5.0),
Joblib (>= 1.1.1),
threadpoolctl (>= 2.0.0).
Build dependencies¶
Building Scikit-learn also requires:
Cython >= 0.29.33
A C/C++ compiler and a matching OpenMP runtime library. See the platform system specific instructions for more details.
Note
If OpenMP is not supported by the compiler, the build will be done with
OpenMP functionalities disabled. This is not recommended since it will force
some estimators to run in sequential mode instead of leveraging thread-based
parallelism. Setting the SKLEARN_FAIL_NO_OPENMP environment variable
(before cythonization) will force the build to fail if OpenMP is not
supported.
Since version 0.21, scikit-learn automatically detects and uses the linear algebra library used by SciPy at runtime. Scikit-learn has therefore no build dependency on BLAS/LAPACK implementations such as OpenBlas, Atlas, Blis or MKL.
Test dependencies¶
Running tests requires:
pytest >= 7.1.2
Some tests also require pandas.
Building a specific version from a tag¶
If you want to build a stable version, you can git checkout <VERSION>
to get the code for that particular version, or download an zip archive of
the version from github.
Editable mode¶
If you run the development version, it is cumbersome to reinstall the package
each time you update the sources. Therefore it is recommended that you install
in with the pip install -v --no-use-pep517 --no-build-isolation -e . command,
which allows you to edit the code in-place. This builds the extension in place and
creates a link to the development directory (see the pip docs).
As the doc above explains, this is fundamentally similar to using the command
python setup.py develop. (see the setuptool docs).
It is however preferred to use pip.
On Unix-like systems, you can equivalently type make in from the top-level
folder. Have a look at the Makefile for additional utilities.
Platform-specific instructions¶
Here are instructions to install a working C/C++ compiler with OpenMP support to build scikit-learn Cython extensions for each supported platform.
Windows¶
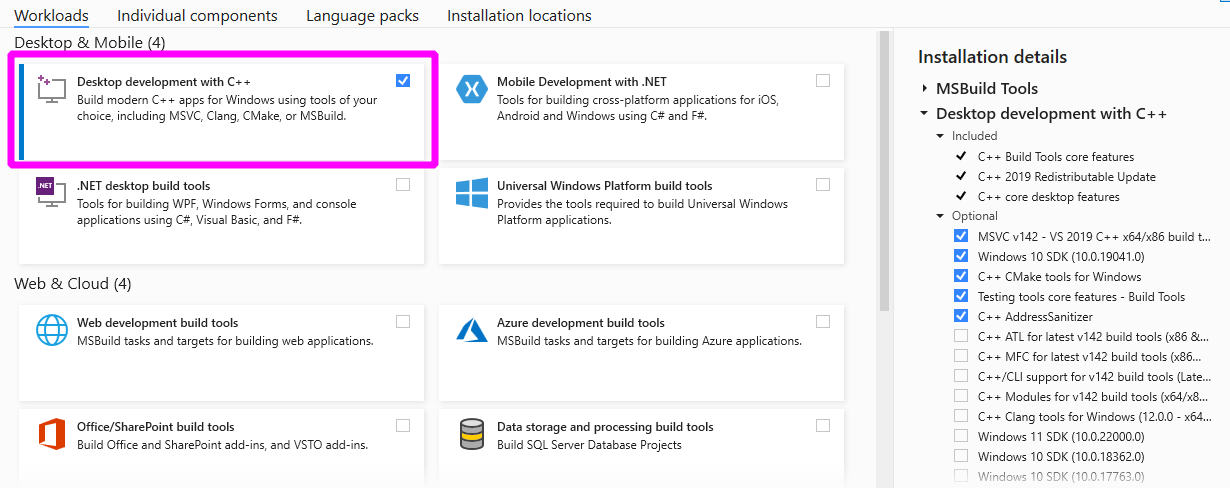
First, download the Build Tools for Visual Studio 2019 installer.
Run the downloaded vs_buildtools.exe file, during the installation you will
need to make sure you select “Desktop development with C++”, similarly to this
screenshot:
Secondly, find out if you are running 64-bit or 32-bit Python. The building
command depends on the architecture of the Python interpreter. You can check
the architecture by running the following in cmd or powershell
console:
python -c "import struct; print(struct.calcsize('P') * 8)"
For 64-bit Python, configure the build environment by running the following
commands in cmd or an Anaconda Prompt (if you use Anaconda):
SET DISTUTILS_USE_SDK=1
"C:\Program Files (x86)\Microsoft Visual Studio\2019\BuildTools\VC\Auxiliary\Build\vcvarsall.bat" x64
Replace x64 by x86 to build for 32-bit Python.
Please be aware that the path above might be different from user to user. The aim is to point to the “vcvarsall.bat” file that will set the necessary environment variables in the current command prompt.
Finally, build scikit-learn from this command prompt:
pip install -v --no-use-pep517 --no-build-isolation -e .
macOS¶
The default C compiler on macOS, Apple clang (confusingly aliased as
/usr/bin/gcc), does not directly support OpenMP. We present two alternatives
to enable OpenMP support:
either install
conda-forge::compilerswith conda;or install
libompwith Homebrew to extend the default Apple clang compiler.
For Apple Silicon M1 hardware, only the conda-forge method below is known to
work at the time of writing (January 2021). You can install the macos/arm64
distribution of conda using the miniforge installer
macOS compilers from conda-forge¶
If you use the conda package manager (version >= 4.7), you can install the
compilers meta-package from the conda-forge channel, which provides
OpenMP-enabled C/C++ compilers based on the llvm toolchain.
First install the macOS command line tools:
xcode-select --install
It is recommended to use a dedicated conda environment to build scikit-learn from source:
conda create -n sklearn-dev -c conda-forge python numpy scipy cython \
joblib threadpoolctl pytest compilers llvm-openmp
It is not always necessary but it is safer to open a new prompt before activating the newly created conda environment.
conda activate sklearn-dev
make clean
pip install -v --no-use-pep517 --no-build-isolation -e .
Note
If you get any conflicting dependency error message, try commenting out
any custom conda configuration in the $HOME/.condarc file. In
particular the channel_priority: strict directive is known to cause
problems for this setup.
You can check that the custom compilers are properly installed from conda forge using the following command:
conda list
which should include compilers and llvm-openmp.
The compilers meta-package will automatically set custom environment variables:
echo $CC
echo $CXX
echo $CFLAGS
echo $CXXFLAGS
echo $LDFLAGS
They point to files and folders from your sklearn-dev conda environment
(in particular in the bin/, include/ and lib/ subfolders). For instance
-L/path/to/conda/envs/sklearn-dev/lib should appear in LDFLAGS.
In the log, you should see the compiled extension being built with the clang
and clang++ compilers installed by conda with the -fopenmp command line
flag.
macOS compilers from Homebrew¶
Another solution is to enable OpenMP support for the clang compiler shipped by default on macOS.
First install the macOS command line tools:
xcode-select --install
Install the Homebrew package manager for macOS.
Install the LLVM OpenMP library:
brew install libomp
Set the following environment variables:
export CC=/usr/bin/clang
export CXX=/usr/bin/clang++
export CPPFLAGS="$CPPFLAGS -Xpreprocessor -fopenmp"
export CFLAGS="$CFLAGS -I/usr/local/opt/libomp/include"
export CXXFLAGS="$CXXFLAGS -I/usr/local/opt/libomp/include"
export LDFLAGS="$LDFLAGS -Wl,-rpath,/usr/local/opt/libomp/lib -L/usr/local/opt/libomp/lib -lomp"
Finally, build scikit-learn in verbose mode (to check for the presence of the
-fopenmp flag in the compiler commands):
make clean
pip install -v --no-use-pep517 --no-build-isolation -e .
Linux¶
Linux compilers from the system¶
Installing scikit-learn from source without using conda requires you to have installed the scikit-learn Python development headers and a working C/C++ compiler with OpenMP support (typically the GCC toolchain).
Install build dependencies for Debian-based operating systems, e.g. Ubuntu:
sudo apt-get install build-essential python3-dev python3-pip
then proceed as usual:
pip3 install cython
pip3 install --verbose --editable .
Cython and the pre-compiled wheels for the runtime dependencies (numpy, scipy
and joblib) should automatically be installed in
$HOME/.local/lib/pythonX.Y/site-packages. Alternatively you can run the
above commands from a virtualenv or a conda environment to get full
isolation from the Python packages installed via the system packager. When
using an isolated environment, pip3 should be replaced by pip in the
above commands.
When precompiled wheels of the runtime dependencies are not available for your architecture (e.g. ARM), you can install the system versions:
sudo apt-get install cython3 python3-numpy python3-scipy
On Red Hat and clones (e.g. CentOS), install the dependencies using:
sudo yum -y install gcc gcc-c++ python3-devel numpy scipy
Linux compilers from conda-forge¶
Alternatively, install a recent version of the GNU C Compiler toolchain (GCC) in the user folder using conda:
conda create -n sklearn-dev -c conda-forge python numpy scipy cython \
joblib threadpoolctl pytest compilers
It is not always necessary but it is safer to open a new prompt before activating the newly created conda environment.
conda activate sklearn-dev
pip install -v --no-use-pep517 --no-build-isolation -e .
FreeBSD¶
The clang compiler included in FreeBSD 12.0 and 11.2 base systems does not
include OpenMP support. You need to install the openmp library from packages
(or ports):
sudo pkg install openmp
This will install header files in /usr/local/include and libs in
/usr/local/lib. Since these directories are not searched by default, you
can set the environment variables to these locations:
export CFLAGS="$CFLAGS -I/usr/local/include"
export CXXFLAGS="$CXXFLAGS -I/usr/local/include"
export LDFLAGS="$LDFLAGS -Wl,-rpath,/usr/local/lib -L/usr/local/lib -lomp"
Finally, build the package using the standard command:
pip install -v --no-use-pep517 --no-build-isolation -e .
For the upcoming FreeBSD 12.1 and 11.3 versions, OpenMP will be included in the base system and these steps will not be necessary.
Alternative compilers¶
The following command will build scikit-learn using your default C/C++ compiler.
pip install --verbose --editable .
If you want to build scikit-learn with another compiler handled by setuptools,
use the following command:
python setup.py build_ext --compiler=<compiler> -i build_clib --compiler=<compiler>
To see the list of available compilers run:
python setup.py build_ext --help-compiler
If your compiler is not listed here, you can specify it through some environment
variables (does not work on windows). This section
of the setuptools documentation explains in details which environment variables
are used by setuptools, and at which stage of the compilation, to set the
compiler and linker options.
When setting these environment variables, it is advised to first check their
sysconfig counterparts variables and adapt them to your compiler. For instance:
import sysconfig
print(sysconfig.get_config_var('CC'))
print(sysconfig.get_config_var('LDFLAGS'))
In addition, since Scikit-learn uses OpenMP, you need to include the appropriate OpenMP
flag of your compiler into the CFLAGS and CPPFLAGS environment variables.
Parallel builds¶
It is possible to build scikit-learn compiled extensions in parallel by setting
and environment variable as follows before calling the pip install or
python setup.py build_ext commands:
export SKLEARN_BUILD_PARALLEL=3
pip install -v --no-use-pep517 --no-build-isolation -e .
On a machine with 2 CPU cores, it can be beneficial to use a parallelism level of 3 to overlap IO bound tasks (reading and writing files on disk) with CPU bound tasks (actually compiling).