Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder
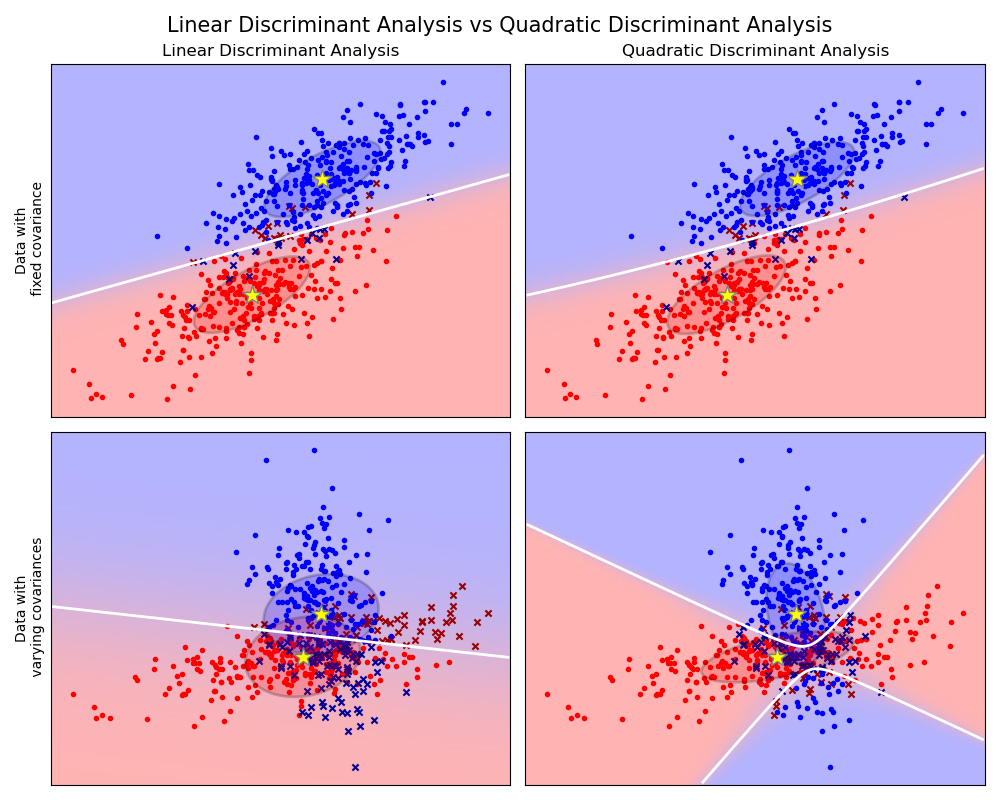
Linear and Quadratic Discriminant Analysis with covariance ellipsoid¶
This example plots the covariance ellipsoids of each class and decision boundary learned by LDA and QDA. The ellipsoids display the double standard deviation for each class. With LDA, the standard deviation is the same for all the classes, while each class has its own standard deviation with QDA.
Colormap¶
import matplotlib as mpl
import matplotlib.pyplot as plt
from matplotlib import colors
cmap = colors.LinearSegmentedColormap(
"red_blue_classes",
{
"red": [(0, 1, 1), (1, 0.7, 0.7)],
"green": [(0, 0.7, 0.7), (1, 0.7, 0.7)],
"blue": [(0, 0.7, 0.7), (1, 1, 1)],
},
)
plt.cm.register_cmap(cmap=cmap)
/home/circleci/project/examples/classification/plot_lda_qda.py:30: MatplotlibDeprecationWarning:
The register_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps.register(name)`` instead.
Datasets generation functions¶
import numpy as np
def dataset_fixed_cov():
"""Generate 2 Gaussians samples with the same covariance matrix"""
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0.0, -0.23], [0.83, 0.23]])
X = np.r_[
np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C) + np.array([1, 1]),
]
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
def dataset_cov():
"""Generate 2 Gaussians samples with different covariance matrices"""
n, dim = 300, 2
np.random.seed(0)
C = np.array([[0.0, -1.0], [2.5, 0.7]]) * 2.0
X = np.r_[
np.dot(np.random.randn(n, dim), C),
np.dot(np.random.randn(n, dim), C.T) + np.array([1, 4]),
]
y = np.hstack((np.zeros(n), np.ones(n)))
return X, y
Plot functions¶
from scipy import linalg
def plot_data(lda, X, y, y_pred, fig_index):
splot = plt.subplot(2, 2, fig_index)
if fig_index == 1:
plt.title("Linear Discriminant Analysis")
plt.ylabel("Data with\n fixed covariance")
elif fig_index == 2:
plt.title("Quadratic Discriminant Analysis")
elif fig_index == 3:
plt.ylabel("Data with\n varying covariances")
tp = y == y_pred # True Positive
tp0, tp1 = tp[y == 0], tp[y == 1]
X0, X1 = X[y == 0], X[y == 1]
X0_tp, X0_fp = X0[tp0], X0[~tp0]
X1_tp, X1_fp = X1[tp1], X1[~tp1]
# class 0: dots
plt.scatter(X0_tp[:, 0], X0_tp[:, 1], marker=".", color="red")
plt.scatter(X0_fp[:, 0], X0_fp[:, 1], marker="x", s=20, color="#990000") # dark red
# class 1: dots
plt.scatter(X1_tp[:, 0], X1_tp[:, 1], marker=".", color="blue")
plt.scatter(
X1_fp[:, 0], X1_fp[:, 1], marker="x", s=20, color="#000099"
) # dark blue
# class 0 and 1 : areas
nx, ny = 200, 100
x_min, x_max = plt.xlim()
y_min, y_max = plt.ylim()
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx), np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.pcolormesh(
xx, yy, Z, cmap="red_blue_classes", norm=colors.Normalize(0.0, 1.0), zorder=0
)
plt.contour(xx, yy, Z, [0.5], linewidths=2.0, colors="white")
# means
plt.plot(
lda.means_[0][0],
lda.means_[0][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
plt.plot(
lda.means_[1][0],
lda.means_[1][1],
"*",
color="yellow",
markersize=15,
markeredgecolor="grey",
)
return splot
def plot_ellipse(splot, mean, cov, color):
v, w = linalg.eigh(cov)
u = w[0] / linalg.norm(w[0])
angle = np.arctan(u[1] / u[0])
angle = 180 * angle / np.pi # convert to degrees
# filled Gaussian at 2 standard deviation
ell = mpl.patches.Ellipse(
mean,
2 * v[0] ** 0.5,
2 * v[1] ** 0.5,
angle=180 + angle,
facecolor=color,
edgecolor="black",
linewidth=2,
)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.2)
splot.add_artist(ell)
splot.set_xticks(())
splot.set_yticks(())
def plot_lda_cov(lda, splot):
plot_ellipse(splot, lda.means_[0], lda.covariance_, "red")
plot_ellipse(splot, lda.means_[1], lda.covariance_, "blue")
def plot_qda_cov(qda, splot):
plot_ellipse(splot, qda.means_[0], qda.covariance_[0], "red")
plot_ellipse(splot, qda.means_[1], qda.covariance_[1], "blue")
Plot¶
plt.figure(figsize=(10, 8), facecolor="white")
plt.suptitle(
"Linear Discriminant Analysis vs Quadratic Discriminant Analysis",
y=0.98,
fontsize=15,
)
from sklearn.discriminant_analysis import (
LinearDiscriminantAnalysis,
QuadraticDiscriminantAnalysis,
)
for i, (X, y) in enumerate([dataset_fixed_cov(), dataset_cov()]):
# Linear Discriminant Analysis
lda = LinearDiscriminantAnalysis(solver="svd", store_covariance=True)
y_pred = lda.fit(X, y).predict(X)
splot = plot_data(lda, X, y, y_pred, fig_index=2 * i + 1)
plot_lda_cov(lda, splot)
plt.axis("tight")
# Quadratic Discriminant Analysis
qda = QuadraticDiscriminantAnalysis(store_covariance=True)
y_pred = qda.fit(X, y).predict(X)
splot = plot_data(qda, X, y, y_pred, fig_index=2 * i + 2)
plot_qda_cov(qda, splot)
plt.axis("tight")
plt.tight_layout()
plt.subplots_adjust(top=0.92)
plt.show()
Total running time of the script: (0 minutes 0.327 seconds)