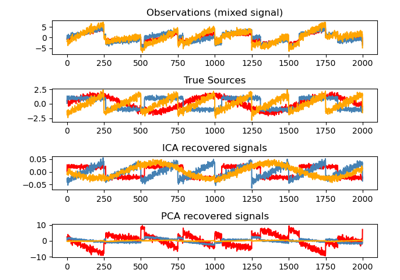
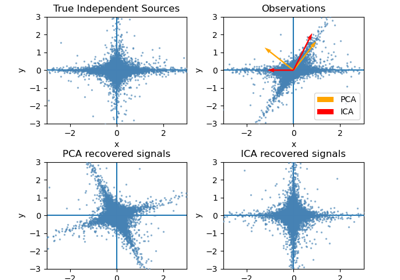
sklearn.decomposition.FastICA¶
- class sklearn.decomposition.FastICA(n_components=None, *, algorithm='parallel', whiten='warn', fun='logcosh', fun_args=None, max_iter=200, tol=0.0001, w_init=None, whiten_solver='svd', random_state=None)[source]¶
FastICA: a fast algorithm for Independent Component Analysis.
The implementation is based on [1].
Read more in the User Guide.
- Parameters:
- n_componentsint, default=None
Number of components to use. If None is passed, all are used.
- algorithm{‘parallel’, ‘deflation’}, default=’parallel’
Specify which algorithm to use for FastICA.
- whitenstr or bool, default=”warn”
Specify the whitening strategy to use.
If ‘arbitrary-variance’ (default), a whitening with variance arbitrary is used.
If ‘unit-variance’, the whitening matrix is rescaled to ensure that each recovered source has unit variance.
If False, the data is already considered to be whitened, and no whitening is performed.
Deprecated since version 1.1: Starting in v1.3,
whiten='unit-variance'will be used by default.whiten=Trueis deprecated from 1.1 and will raise ValueError in 1.3. Usewhiten=arbitrary-varianceinstead.- fun{‘logcosh’, ‘exp’, ‘cube’} or callable, default=’logcosh’
The functional form of the G function used in the approximation to neg-entropy. Could be either ‘logcosh’, ‘exp’, or ‘cube’. You can also provide your own function. It should return a tuple containing the value of the function, and of its derivative, in the point. The derivative should be averaged along its last dimension. Example:
def my_g(x): return x ** 3, (3 * x ** 2).mean(axis=-1)
- fun_argsdict, default=None
Arguments to send to the functional form. If empty or None and if fun=’logcosh’, fun_args will take value {‘alpha’ : 1.0}.
- max_iterint, default=200
Maximum number of iterations during fit.
- tolfloat, default=1e-4
A positive scalar giving the tolerance at which the un-mixing matrix is considered to have converged.
- w_initarray-like of shape (n_components, n_components), default=None
Initial un-mixing array. If
w_init=None, then an array of values drawn from a normal distribution is used.- whiten_solver{“eigh”, “svd”}, default=”svd”
The solver to use for whitening.
“svd” is more stable numerically if the problem is degenerate, and often faster when
n_samples <= n_features.“eigh” is generally more memory efficient when
n_samples >= n_features, and can be faster whenn_samples >= 50 * n_features.
New in version 1.2.
- random_stateint, RandomState instance or None, default=None
Used to initialize
w_initwhen not specified, with a normal distribution. Pass an int, for reproducible results across multiple function calls. See Glossary.
- Attributes:
- components_ndarray of shape (n_components, n_features)
The linear operator to apply to the data to get the independent sources. This is equal to the unmixing matrix when
whitenis False, and equal tonp.dot(unmixing_matrix, self.whitening_)whenwhitenis True.- mixing_ndarray of shape (n_features, n_components)
The pseudo-inverse of
components_. It is the linear operator that maps independent sources to the data.- mean_ndarray of shape(n_features,)
The mean over features. Only set if
self.whitenis True.- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
- n_iter_int
If the algorithm is “deflation”, n_iter is the maximum number of iterations run across all components. Else they are just the number of iterations taken to converge.
- whitening_ndarray of shape (n_components, n_features)
Only set if whiten is ‘True’. This is the pre-whitening matrix that projects data onto the first
n_componentsprincipal components.
See also
PCAPrincipal component analysis (PCA).
IncrementalPCAIncremental principal components analysis (IPCA).
KernelPCAKernel Principal component analysis (KPCA).
MiniBatchSparsePCAMini-batch Sparse Principal Components Analysis.
SparsePCASparse Principal Components Analysis (SparsePCA).
References
[1]A. Hyvarinen and E. Oja, Independent Component Analysis: Algorithms and Applications, Neural Networks, 13(4-5), 2000, pp. 411-430.
Examples
>>> from sklearn.datasets import load_digits >>> from sklearn.decomposition import FastICA >>> X, _ = load_digits(return_X_y=True) >>> transformer = FastICA(n_components=7, ... random_state=0, ... whiten='unit-variance') >>> X_transformed = transformer.fit_transform(X) >>> X_transformed.shape (1797, 7)
Methods
fit(X[, y])Fit the model to X.
fit_transform(X[, y])Fit the model and recover the sources from X.
get_feature_names_out([input_features])Get output feature names for transformation.
get_params([deep])Get parameters for this estimator.
inverse_transform(X[, copy])Transform the sources back to the mixed data (apply mixing matrix).
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X[, copy])Recover the sources from X (apply the unmixing matrix).
- fit(X, y=None)[source]¶
Fit the model to X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Training data, where
n_samplesis the number of samples andn_featuresis the number of features.- yIgnored
Not used, present for API consistency by convention.
- Returns:
- selfobject
Returns the instance itself.
- fit_transform(X, y=None)[source]¶
Fit the model and recover the sources from X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Training data, where
n_samplesis the number of samples andn_featuresis the number of features.- yIgnored
Not used, present for API consistency by convention.
- Returns:
- X_newndarray of shape (n_samples, n_components)
Estimated sources obtained by transforming the data with the estimated unmixing matrix.
- get_feature_names_out(input_features=None)[source]¶
Get output feature names for transformation.
The feature names out will prefixed by the lowercased class name. For example, if the transformer outputs 3 features, then the feature names out are:
["class_name0", "class_name1", "class_name2"].- Parameters:
- input_featuresarray-like of str or None, default=None
Only used to validate feature names with the names seen in
fit.
- Returns:
- feature_names_outndarray of str objects
Transformed feature names.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- inverse_transform(X, copy=True)[source]¶
Transform the sources back to the mixed data (apply mixing matrix).
- Parameters:
- Xarray-like of shape (n_samples, n_components)
Sources, where
n_samplesis the number of samples andn_componentsis the number of components.- copybool, default=True
If False, data passed to fit are overwritten. Defaults to True.
- Returns:
- X_newndarray of shape (n_samples, n_features)
Reconstructed data obtained with the mixing matrix.
- set_output(*, transform=None)[source]¶
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”}, default=None
Configure output of
transformandfit_transform."default": Default output format of a transformer"pandas": DataFrame outputNone: Transform configuration is unchanged
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- transform(X, copy=True)[source]¶
Recover the sources from X (apply the unmixing matrix).
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Data to transform, where
n_samplesis the number of samples andn_featuresis the number of features.- copybool, default=True
If False, data passed to fit can be overwritten. Defaults to True.
- Returns:
- X_newndarray of shape (n_samples, n_components)
Estimated sources obtained by transforming the data with the estimated unmixing matrix.