sklearn.preprocessing.MinMaxScaler¶
- class sklearn.preprocessing.MinMaxScaler(feature_range=(0, 1), *, copy=True, clip=False)[source]¶
Transform features by scaling each feature to a given range.
This estimator scales and translates each feature individually such that it is in the given range on the training set, e.g. between zero and one.
The transformation is given by:
X_std = (X - X.min(axis=0)) / (X.max(axis=0) - X.min(axis=0)) X_scaled = X_std * (max - min) + min
where min, max = feature_range.
This transformation is often used as an alternative to zero mean, unit variance scaling.
Read more in the User Guide.
- Parameters:
- feature_rangetuple (min, max), default=(0, 1)
Desired range of transformed data.
- copybool, default=True
Set to False to perform inplace row normalization and avoid a copy (if the input is already a numpy array).
- clipbool, default=False
Set to True to clip transformed values of held-out data to provided
feature range.New in version 0.24.
- Attributes:
- min_ndarray of shape (n_features,)
Per feature adjustment for minimum. Equivalent to
min - X.min(axis=0) * self.scale_- scale_ndarray of shape (n_features,)
Per feature relative scaling of the data. Equivalent to
(max - min) / (X.max(axis=0) - X.min(axis=0))New in version 0.17: scale_ attribute.
- data_min_ndarray of shape (n_features,)
Per feature minimum seen in the data
New in version 0.17: data_min_
- data_max_ndarray of shape (n_features,)
Per feature maximum seen in the data
New in version 0.17: data_max_
- data_range_ndarray of shape (n_features,)
Per feature range
(data_max_ - data_min_)seen in the dataNew in version 0.17: data_range_
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- n_samples_seen_int
The number of samples processed by the estimator. It will be reset on new calls to fit, but increments across
partial_fitcalls.- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
minmax_scaleEquivalent function without the estimator API.
Notes
NaNs are treated as missing values: disregarded in fit, and maintained in transform.
For a comparison of the different scalers, transformers, and normalizers, see examples/preprocessing/plot_all_scaling.py.
Examples
>>> from sklearn.preprocessing import MinMaxScaler >>> data = [[-1, 2], [-0.5, 6], [0, 10], [1, 18]] >>> scaler = MinMaxScaler() >>> print(scaler.fit(data)) MinMaxScaler() >>> print(scaler.data_max_) [ 1. 18.] >>> print(scaler.transform(data)) [[0. 0. ] [0.25 0.25] [0.5 0.5 ] [1. 1. ]] >>> print(scaler.transform([[2, 2]])) [[1.5 0. ]]
Methods
fit(X[, y])Compute the minimum and maximum to be used for later scaling.
fit_transform(X[, y])Fit to data, then transform it.
get_feature_names_out([input_features])Get output feature names for transformation.
get_params([deep])Get parameters for this estimator.
Undo the scaling of X according to feature_range.
partial_fit(X[, y])Online computation of min and max on X for later scaling.
set_params(**params)Set the parameters of this estimator.
transform(X)Scale features of X according to feature_range.
- fit(X, y=None)[source]¶
Compute the minimum and maximum to be used for later scaling.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
The data used to compute the per-feature minimum and maximum used for later scaling along the features axis.
- yNone
Ignored.
- Returns:
- selfobject
Fitted scaler.
- fit_transform(X, y=None, **fit_params)[source]¶
Fit to data, then transform it.
Fits transformer to
Xandywith optional parametersfit_paramsand returns a transformed version ofX.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs), default=None
Target values (None for unsupervised transformations).
- **fit_paramsdict
Additional fit parameters.
- Returns:
- X_newndarray array of shape (n_samples, n_features_new)
Transformed array.
- get_feature_names_out(input_features=None)[source]¶
Get output feature names for transformation.
- Parameters:
- input_featuresarray-like of str or None, default=None
Input features.
If
input_featuresisNone, thenfeature_names_in_is used as feature names in. Iffeature_names_in_is not defined, then the following input feature names are generated:["x0", "x1", ..., "x(n_features_in_ - 1)"].If
input_featuresis an array-like, theninput_featuresmust matchfeature_names_in_iffeature_names_in_is defined.
- Returns:
- feature_names_outndarray of str objects
Same as input features.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- inverse_transform(X)[source]¶
Undo the scaling of X according to feature_range.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input data that will be transformed. It cannot be sparse.
- Returns:
- Xtndarray of shape (n_samples, n_features)
Transformed data.
- partial_fit(X, y=None)[source]¶
Online computation of min and max on X for later scaling.
All of X is processed as a single batch. This is intended for cases when
fitis not feasible due to very large number ofn_samplesor because X is read from a continuous stream.- Parameters:
- Xarray-like of shape (n_samples, n_features)
The data used to compute the mean and standard deviation used for later scaling along the features axis.
- yNone
Ignored.
- Returns:
- selfobject
Fitted scaler.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using sklearn.preprocessing.MinMaxScaler¶
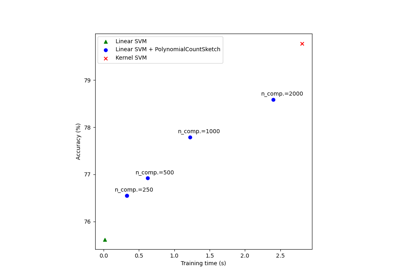
Scalable learning with polynomial kernel approximation
Manifold learning on handwritten digits: Locally Linear Embedding, Isomap…
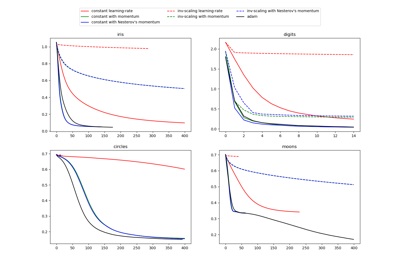
Compare Stochastic learning strategies for MLPClassifier
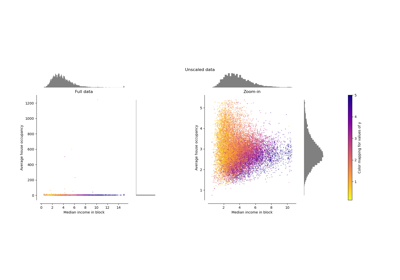
Compare the effect of different scalers on data with outliers