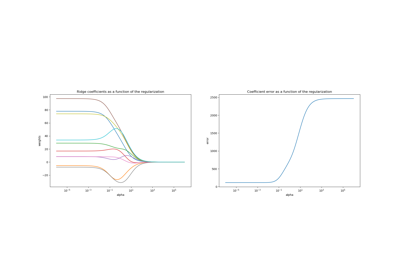
sklearn.linear_model.Ridge¶
-
class
sklearn.linear_model.Ridge(alpha=1.0, fit_intercept=True, normalize=False, copy_X=True, max_iter=None, tol=0.001, solver=’auto’, random_state=None)[source]¶ Linear least squares with l2 regularization.
Minimizes the objective function:
||y - Xw||^2_2 + alpha * ||w||^2_2
This model solves a regression model where the loss function is the linear least squares function and regularization is given by the l2-norm. Also known as Ridge Regression or Tikhonov regularization. This estimator has built-in support for multi-variate regression (i.e., when y is a 2d-array of shape [n_samples, n_targets]).
Read more in the User Guide.
Parameters: - alpha : {float, array-like}, shape (n_targets)
Regularization strength; must be a positive float. Regularization improves the conditioning of the problem and reduces the variance of the estimates. Larger values specify stronger regularization. Alpha corresponds to
C^-1in other linear models such as LogisticRegression or LinearSVC. If an array is passed, penalties are assumed to be specific to the targets. Hence they must correspond in number.- fit_intercept : boolean
Whether to calculate the intercept for this model. If set to false, no intercept will be used in calculations (e.g. data is expected to be already centered).
- normalize : boolean, optional, default False
This parameter is ignored when
fit_interceptis set to False. If True, the regressors X will be normalized before regression by subtracting the mean and dividing by the l2-norm. If you wish to standardize, please usesklearn.preprocessing.StandardScalerbefore callingfiton an estimator withnormalize=False.- copy_X : boolean, optional, default True
If True, X will be copied; else, it may be overwritten.
- max_iter : int, optional
Maximum number of iterations for conjugate gradient solver. For ‘sparse_cg’ and ‘lsqr’ solvers, the default value is determined by scipy.sparse.linalg. For ‘sag’ solver, the default value is 1000.
- tol : float
Precision of the solution.
- solver : {‘auto’, ‘svd’, ‘cholesky’, ‘lsqr’, ‘sparse_cg’, ‘sag’, ‘saga’}
Solver to use in the computational routines:
- ‘auto’ chooses the solver automatically based on the type of data.
- ‘svd’ uses a Singular Value Decomposition of X to compute the Ridge coefficients. More stable for singular matrices than ‘cholesky’.
- ‘cholesky’ uses the standard scipy.linalg.solve function to obtain a closed-form solution.
- ‘sparse_cg’ uses the conjugate gradient solver as found in
scipy.sparse.linalg.cg. As an iterative algorithm, this solver is
more appropriate than ‘cholesky’ for large-scale data
(possibility to set
toland max_iter). - ‘lsqr’ uses the dedicated regularized least-squares routine scipy.sparse.linalg.lsqr. It is the fastest and uses an iterative procedure.
- ‘sag’ uses a Stochastic Average Gradient descent, and ‘saga’ uses its improved, unbiased version named SAGA. Both methods also use an iterative procedure, and are often faster than other solvers when both n_samples and n_features are large. Note that ‘sag’ and ‘saga’ fast convergence is only guaranteed on features with approximately the same scale. You can preprocess the data with a scaler from sklearn.preprocessing.
All last five solvers support both dense and sparse data. However, only ‘sag’ and ‘sparse_cg’ supports sparse input when
fit_interceptis True.New in version 0.17: Stochastic Average Gradient descent solver.
New in version 0.19: SAGA solver.
- random_state : int, RandomState instance or None, optional, default None
The seed of the pseudo random number generator to use when shuffling the data. If int, random_state is the seed used by the random number generator; If RandomState instance, random_state is the random number generator; If None, the random number generator is the RandomState instance used by
np.random. Used whensolver== ‘sag’.New in version 0.17: random_state to support Stochastic Average Gradient.
Attributes: - coef_ : array, shape (n_features,) or (n_targets, n_features)
Weight vector(s).
- intercept_ : float | array, shape = (n_targets,)
Independent term in decision function. Set to 0.0 if
fit_intercept = False.- n_iter_ : array or None, shape (n_targets,)
Actual number of iterations for each target. Available only for sag and lsqr solvers. Other solvers will return None.
New in version 0.17.
See also
RidgeClassifier- Ridge classifier
RidgeCV- Ridge regression with built-in cross validation
sklearn.kernel_ridge.KernelRidge- Kernel ridge regression combines ridge regression with the kernel trick
Examples
>>> from sklearn.linear_model import Ridge >>> import numpy as np >>> n_samples, n_features = 10, 5 >>> rng = np.random.RandomState(0) >>> y = rng.randn(n_samples) >>> X = rng.randn(n_samples, n_features) >>> clf = Ridge(alpha=1.0) >>> clf.fit(X, y) Ridge(alpha=1.0, copy_X=True, fit_intercept=True, max_iter=None, normalize=False, random_state=None, solver='auto', tol=0.001)
Methods
fit(self, X, y[, sample_weight])Fit Ridge regression model get_params(self[, deep])Get parameters for this estimator. predict(self, X)Predict using the linear model score(self, X, y[, sample_weight])Returns the coefficient of determination R^2 of the prediction. set_params(self, \*\*params)Set the parameters of this estimator. -
__init__(self, alpha=1.0, fit_intercept=True, normalize=False, copy_X=True, max_iter=None, tol=0.001, solver=’auto’, random_state=None)[source]¶
-
fit(self, X, y, sample_weight=None)[source]¶ Fit Ridge regression model
Parameters: - X : {array-like, sparse matrix}, shape = [n_samples, n_features]
Training data
- y : array-like, shape = [n_samples] or [n_samples, n_targets]
Target values
- sample_weight : float or numpy array of shape [n_samples]
Individual weights for each sample
Returns: - self : returns an instance of self.
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
predict(self, X)[source]¶ Predict using the linear model
Parameters: - X : array_like or sparse matrix, shape (n_samples, n_features)
Samples.
Returns: - C : array, shape (n_samples,)
Returns predicted values.
-
score(self, X, y, sample_weight=None)[source]¶ Returns the coefficient of determination R^2 of the prediction.
The coefficient R^2 is defined as (1 - u/v), where u is the residual sum of squares ((y_true - y_pred) ** 2).sum() and v is the total sum of squares ((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a R^2 score of 0.0.
Parameters: - X : array-like, shape = (n_samples, n_features)
Test samples. For some estimators this may be a precomputed kernel matrix instead, shape = (n_samples, n_samples_fitted], where n_samples_fitted is the number of samples used in the fitting for the estimator.
- y : array-like, shape = (n_samples) or (n_samples, n_outputs)
True values for X.
- sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns: - score : float
R^2 of self.predict(X) wrt. y.
Notes
The R2 score used when calling
scoreon a regressor will usemultioutput='uniform_average'from version 0.23 to keep consistent withmetrics.r2_score. This will influence thescoremethod of all the multioutput regressors (except formultioutput.MultiOutputRegressor). To specify the default value manually and avoid the warning, please either callmetrics.r2_scoredirectly or make a custom scorer withmetrics.make_scorer(the built-in scorer'r2'usesmultioutput='uniform_average').
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self