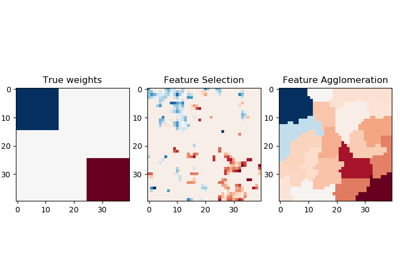
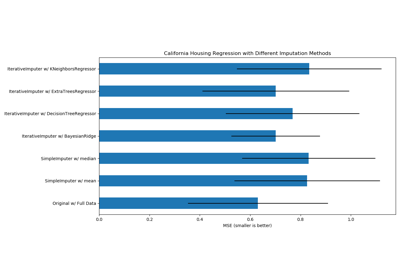
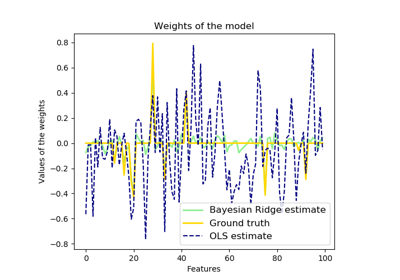
sklearn.linear_model.BayesianRidge¶
-
class
sklearn.linear_model.BayesianRidge(n_iter=300, tol=0.001, alpha_1=1e-06, alpha_2=1e-06, lambda_1=1e-06, lambda_2=1e-06, compute_score=False, fit_intercept=True, normalize=False, copy_X=True, verbose=False)[source]¶ Bayesian ridge regression.
Fit a Bayesian ridge model. See the Notes section for details on this implementation and the optimization of the regularization parameters lambda (precision of the weights) and alpha (precision of the noise).
Read more in the User Guide.
Parameters: - n_iter : int, optional
Maximum number of iterations. Default is 300. Should be greater than or equal to 1.
- tol : float, optional
Stop the algorithm if w has converged. Default is 1.e-3.
- alpha_1 : float, optional
Hyper-parameter : shape parameter for the Gamma distribution prior over the alpha parameter. Default is 1.e-6
- alpha_2 : float, optional
Hyper-parameter : inverse scale parameter (rate parameter) for the Gamma distribution prior over the alpha parameter. Default is 1.e-6.
- lambda_1 : float, optional
Hyper-parameter : shape parameter for the Gamma distribution prior over the lambda parameter. Default is 1.e-6.
- lambda_2 : float, optional
Hyper-parameter : inverse scale parameter (rate parameter) for the Gamma distribution prior over the lambda parameter. Default is 1.e-6
- compute_score : boolean, optional
If True, compute the log marginal likelihood at each iteration of the optimization. Default is False.
- fit_intercept : boolean, optional, default True
Whether to calculate the intercept for this model. The intercept is not treated as a probabilistic parameter and thus has no associated variance. If set to False, no intercept will be used in calculations (e.g. data is expected to be already centered).
- normalize : boolean, optional, default False
This parameter is ignored when
fit_interceptis set to False. If True, the regressors X will be normalized before regression by subtracting the mean and dividing by the l2-norm. If you wish to standardize, please usesklearn.preprocessing.StandardScalerbefore callingfiton an estimator withnormalize=False.- copy_X : boolean, optional, default True
If True, X will be copied; else, it may be overwritten.
- verbose : boolean, optional, default False
Verbose mode when fitting the model.
Attributes: - coef_ : array, shape = (n_features,)
Coefficients of the regression model (mean of distribution).
- intercept_ : float
Independent term in decision function. Set to 0.0 if
fit_intercept = False.- alpha_ : float
Estimated precision of the noise.
- lambda_ : float
Estimated precision of the weights.
- sigma_ : array, shape = (n_features, n_features)
Estimated variance-covariance matrix of the weights.
- scores_ : array, shape = (n_iter_ + 1,)
If computed_score is True, value of the log marginal likelihood (to be maximized) at each iteration of the optimization. The array starts with the value of the log marginal likelihood obtained for the initial values of alpha and lambda and ends with the value obtained for the estimated alpha and lambda.
- n_iter_ : int
The actual number of iterations to reach the stopping criterion.
Notes
There exist several strategies to perform Bayesian ridge regression. This implementation is based on the algorithm described in Appendix A of (Tipping, 2001) where updates of the regularization parameters are done as suggested in (MacKay, 1992). Note that according to A New View of Automatic Relevance Determination (Wipf and Nagarajan, 2008) these update rules do not guarantee that the marginal likelihood is increasing between two consecutive iterations of the optimization.
References
D. J. C. MacKay, Bayesian Interpolation, Computation and Neural Systems, Vol. 4, No. 3, 1992.
M. E. Tipping, Sparse Bayesian Learning and the Relevance Vector Machine, Journal of Machine Learning Research, Vol. 1, 2001.
Examples
>>> from sklearn import linear_model >>> clf = linear_model.BayesianRidge() >>> clf.fit([[0,0], [1, 1], [2, 2]], [0, 1, 2]) ... BayesianRidge(alpha_1=1e-06, alpha_2=1e-06, compute_score=False, copy_X=True, fit_intercept=True, lambda_1=1e-06, lambda_2=1e-06, n_iter=300, normalize=False, tol=0.001, verbose=False) >>> clf.predict([[1, 1]]) array([1.])
Methods
fit(self, X, y[, sample_weight])Fit the model get_params(self[, deep])Get parameters for this estimator. predict(self, X[, return_std])Predict using the linear model. score(self, X, y[, sample_weight])Returns the coefficient of determination R^2 of the prediction. set_params(self, \*\*params)Set the parameters of this estimator. -
__init__(self, n_iter=300, tol=0.001, alpha_1=1e-06, alpha_2=1e-06, lambda_1=1e-06, lambda_2=1e-06, compute_score=False, fit_intercept=True, normalize=False, copy_X=True, verbose=False)[source]¶
-
fit(self, X, y, sample_weight=None)[source]¶ Fit the model
Parameters: - X : numpy array of shape [n_samples,n_features]
Training data
- y : numpy array of shape [n_samples]
Target values. Will be cast to X’s dtype if necessary
- sample_weight : numpy array of shape [n_samples]
Individual weights for each sample
New in version 0.20: parameter sample_weight support to BayesianRidge.
Returns: - self : returns an instance of self.
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
predict(self, X, return_std=False)[source]¶ Predict using the linear model.
In addition to the mean of the predictive distribution, also its standard deviation can be returned.
Parameters: - X : {array-like, sparse matrix}, shape = (n_samples, n_features)
Samples.
- return_std : boolean, optional
Whether to return the standard deviation of posterior prediction.
Returns: - y_mean : array, shape = (n_samples,)
Mean of predictive distribution of query points.
- y_std : array, shape = (n_samples,)
Standard deviation of predictive distribution of query points.
-
score(self, X, y, sample_weight=None)[source]¶ Returns the coefficient of determination R^2 of the prediction.
The coefficient R^2 is defined as (1 - u/v), where u is the residual sum of squares ((y_true - y_pred) ** 2).sum() and v is the total sum of squares ((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a R^2 score of 0.0.
Parameters: - X : array-like, shape = (n_samples, n_features)
Test samples. For some estimators this may be a precomputed kernel matrix instead, shape = (n_samples, n_samples_fitted], where n_samples_fitted is the number of samples used in the fitting for the estimator.
- y : array-like, shape = (n_samples) or (n_samples, n_outputs)
True values for X.
- sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns: - score : float
R^2 of self.predict(X) wrt. y.
Notes
The R2 score used when calling
scoreon a regressor will usemultioutput='uniform_average'from version 0.23 to keep consistent withmetrics.r2_score. This will influence thescoremethod of all the multioutput regressors (except formultioutput.MultiOutputRegressor). To specify the default value manually and avoid the warning, please either callmetrics.r2_scoredirectly or make a custom scorer withmetrics.make_scorer(the built-in scorer'r2'usesmultioutput='uniform_average').
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self