Note
Click here to download the full example code
Robust linear model estimation using RANSAC¶
In this example we see how to robustly fit a linear model to faulty data using the RANSAC algorithm.
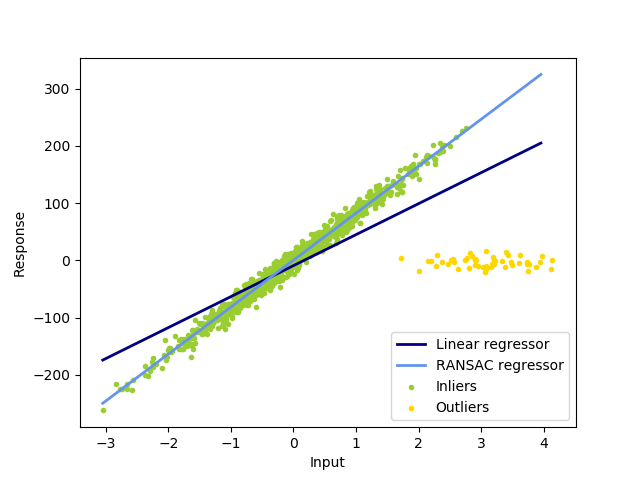
Out:
Estimated coefficients (true, linear regression, RANSAC):
82.1903908407869 [54.17236387] [82.08533159]
import numpy as np
from matplotlib import pyplot as plt
from sklearn import linear_model, datasets
n_samples = 1000
n_outliers = 50
X, y, coef = datasets.make_regression(n_samples=n_samples, n_features=1,
n_informative=1, noise=10,
coef=True, random_state=0)
# Add outlier data
np.random.seed(0)
X[:n_outliers] = 3 + 0.5 * np.random.normal(size=(n_outliers, 1))
y[:n_outliers] = -3 + 10 * np.random.normal(size=n_outliers)
# Fit line using all data
lr = linear_model.LinearRegression()
lr.fit(X, y)
# Robustly fit linear model with RANSAC algorithm
ransac = linear_model.RANSACRegressor()
ransac.fit(X, y)
inlier_mask = ransac.inlier_mask_
outlier_mask = np.logical_not(inlier_mask)
# Predict data of estimated models
line_X = np.arange(X.min(), X.max())[:, np.newaxis]
line_y = lr.predict(line_X)
line_y_ransac = ransac.predict(line_X)
# Compare estimated coefficients
print("Estimated coefficients (true, linear regression, RANSAC):")
print(coef, lr.coef_, ransac.estimator_.coef_)
lw = 2
plt.scatter(X[inlier_mask], y[inlier_mask], color='yellowgreen', marker='.',
label='Inliers')
plt.scatter(X[outlier_mask], y[outlier_mask], color='gold', marker='.',
label='Outliers')
plt.plot(line_X, line_y, color='navy', linewidth=lw, label='Linear regressor')
plt.plot(line_X, line_y_ransac, color='cornflowerblue', linewidth=lw,
label='RANSAC regressor')
plt.legend(loc='lower right')
plt.xlabel("Input")
plt.ylabel("Response")
plt.show()
Total running time of the script: ( 0 minutes 0.285 seconds)

