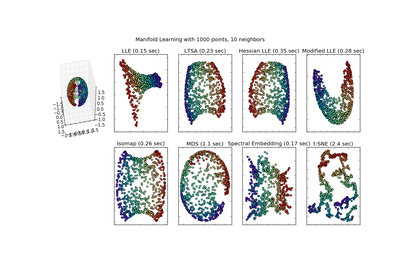
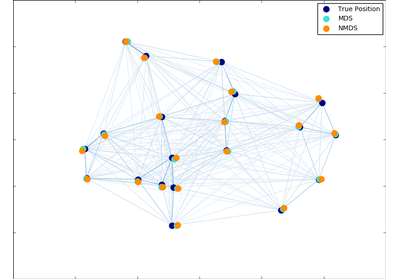
sklearn.manifold.MDS¶
-
class
sklearn.manifold.MDS(n_components=2, metric=True, n_init=4, max_iter=300, verbose=0, eps=0.001, n_jobs=1, random_state=None, dissimilarity='euclidean')[source]¶ Multidimensional scaling
Read more in the User Guide.
Parameters: metric : boolean, optional, default: True
compute metric or nonmetric SMACOF (Scaling by Majorizing a Complicated Function) algorithm
n_components : int, optional, default: 2
number of dimension in which to immerse the similarities overridden if initial array is provided.
n_init : int, optional, default: 4
Number of time the smacof algorithm will be run with different initialisation. The final results will be the best output of the n_init consecutive runs in terms of stress.
max_iter : int, optional, default: 300
Maximum number of iterations of the SMACOF algorithm for a single run
verbose : int, optional, default: 0
level of verbosity
eps : float, optional, default: 1e-6
relative tolerance w.r.t stress to declare converge
n_jobs : int, optional, default: 1
The number of jobs to use for the computation. This works by breaking down the pairwise matrix into n_jobs even slices and computing them in parallel.
If -1 all CPUs are used. If 1 is given, no parallel computing code is used at all, which is useful for debugging. For n_jobs below -1, (n_cpus + 1 + n_jobs) are used. Thus for n_jobs = -2, all CPUs but one are used.
random_state : integer or numpy.RandomState, optional
The generator used to initialize the centers. If an integer is given, it fixes the seed. Defaults to the global numpy random number generator.
dissimilarity : string
Which dissimilarity measure to use. Supported are ‘euclidean’ and ‘precomputed’.
Attributes: embedding_ : array-like, shape [n_components, n_samples]
Stores the position of the dataset in the embedding space
stress_ : float
The final value of the stress (sum of squared distance of the disparities and the distances for all constrained points)
References
“Modern Multidimensional Scaling - Theory and Applications” Borg, I.; Groenen P. Springer Series in Statistics (1997)
“Nonmetric multidimensional scaling: a numerical method” Kruskal, J. Psychometrika, 29 (1964)
“Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis” Kruskal, J. Psychometrika, 29, (1964)
Methods
fit(X[, y, init])Computes the position of the points in the embedding space fit_transform(X[, y, init])Fit the data from X, and returns the embedded coordinates get_params([deep])Get parameters for this estimator. set_params(\*\*params)Set the parameters of this estimator. -
__init__(n_components=2, metric=True, n_init=4, max_iter=300, verbose=0, eps=0.001, n_jobs=1, random_state=None, dissimilarity='euclidean')[source]¶
-
fit(X, y=None, init=None)[source]¶ Computes the position of the points in the embedding space
Parameters: X : array, shape=[n_samples, n_features], or [n_samples, n_samples] if dissimilarity=’precomputed’
Input data.
init : {None or ndarray, shape (n_samples,)}, optional
If None, randomly chooses the initial configuration if ndarray, initialize the SMACOF algorithm with this array.
-
fit_transform(X, y=None, init=None)[source]¶ Fit the data from X, and returns the embedded coordinates
Parameters: X : array, shape=[n_samples, n_features], or [n_samples, n_samples] if dissimilarity=’precomputed’
Input data.
init : {None or ndarray, shape (n_samples,)}, optional
If None, randomly chooses the initial configuration if ndarray, initialize the SMACOF algorithm with this array.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
Parameters: deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: self :
-