Robust Scaling on Toy Data¶
Making sure that each Feature has approximately the same scale can be a
crucial preprocessing step. However, when data contains outliers,
StandardScaler can often
be mislead. In such cases, it is better to use a scaler that is robust
against outliers.
Here, we demonstrate this on a toy dataset, where one single datapoint is a large outlier.
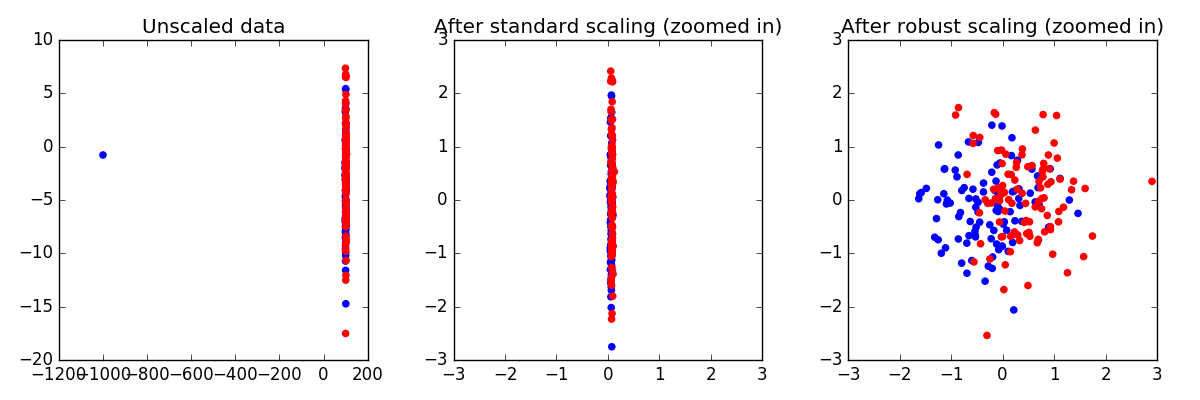
Out:
Testset accuracy using standard scaler: 0.545
Testset accuracy using robust scaler: 0.705
from __future__ import print_function
print(__doc__)
# Code source: Thomas Unterthiner
# License: BSD 3 clause
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import StandardScaler, RobustScaler
# Create training and test data
np.random.seed(42)
n_datapoints = 100
Cov = [[0.9, 0.0], [0.0, 20.0]]
mu1 = [100.0, -3.0]
mu2 = [101.0, -3.0]
X1 = np.random.multivariate_normal(mean=mu1, cov=Cov, size=n_datapoints)
X2 = np.random.multivariate_normal(mean=mu2, cov=Cov, size=n_datapoints)
Y_train = np.hstack([[-1]*n_datapoints, [1]*n_datapoints])
X_train = np.vstack([X1, X2])
X1 = np.random.multivariate_normal(mean=mu1, cov=Cov, size=n_datapoints)
X2 = np.random.multivariate_normal(mean=mu2, cov=Cov, size=n_datapoints)
Y_test = np.hstack([[-1]*n_datapoints, [1]*n_datapoints])
X_test = np.vstack([X1, X2])
X_train[0, 0] = -1000 # a fairly large outlier
# Scale data
standard_scaler = StandardScaler()
Xtr_s = standard_scaler.fit_transform(X_train)
Xte_s = standard_scaler.transform(X_test)
robust_scaler = RobustScaler()
Xtr_r = robust_scaler.fit_transform(X_train)
Xte_r = robust_scaler.transform(X_test)
# Plot data
fig, ax = plt.subplots(1, 3, figsize=(12, 4))
ax[0].scatter(X_train[:, 0], X_train[:, 1],
color=np.where(Y_train > 0, 'r', 'b'))
ax[1].scatter(Xtr_s[:, 0], Xtr_s[:, 1], color=np.where(Y_train > 0, 'r', 'b'))
ax[2].scatter(Xtr_r[:, 0], Xtr_r[:, 1], color=np.where(Y_train > 0, 'r', 'b'))
ax[0].set_title("Unscaled data")
ax[1].set_title("After standard scaling (zoomed in)")
ax[2].set_title("After robust scaling (zoomed in)")
# for the scaled data, we zoom in to the data center (outlier can't be seen!)
for a in ax[1:]:
a.set_xlim(-3, 3)
a.set_ylim(-3, 3)
plt.tight_layout()
plt.show()
# Classify using k-NN
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier()
knn.fit(Xtr_s, Y_train)
acc_s = knn.score(Xte_s, Y_test)
print("Testset accuracy using standard scaler: %.3f" % acc_s)
knn.fit(Xtr_r, Y_train)
acc_r = knn.score(Xte_r, Y_test)
print("Testset accuracy using robust scaler: %.3f" % acc_r)
Total running time of the script: (0 minutes 0.284 seconds)
Download Python source code:
plot_robust_scaling.py
Download IPython notebook:
plot_robust_scaling.ipynb
