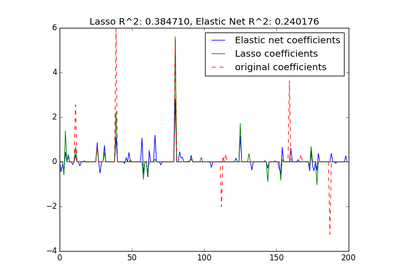
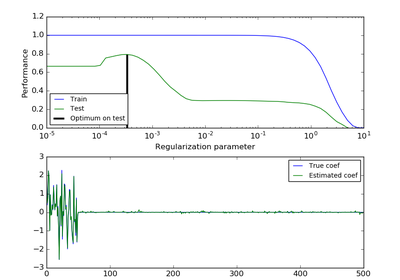
sklearn.linear_model.ElasticNet¶
-
class
sklearn.linear_model.ElasticNet(alpha=1.0, l1_ratio=0.5, fit_intercept=True, normalize=False, precompute=False, max_iter=1000, copy_X=True, tol=0.0001, warm_start=False, positive=False, random_state=None, selection='cyclic')[source]¶ Linear regression with combined L1 and L2 priors as regularizer.
Minimizes the objective function:
1 / (2 * n_samples) * ||y - Xw||^2_2 + + alpha * l1_ratio * ||w||_1 + 0.5 * alpha * (1 - l1_ratio) * ||w||^2_2
If you are interested in controlling the L1 and L2 penalty separately, keep in mind that this is equivalent to:
a * L1 + b * L2
where:
alpha = a + b and l1_ratio = a / (a + b)
The parameter l1_ratio corresponds to alpha in the glmnet R package while alpha corresponds to the lambda parameter in glmnet. Specifically, l1_ratio = 1 is the lasso penalty. Currently, l1_ratio <= 0.01 is not reliable, unless you supply your own sequence of alpha.
Read more in the User Guide.
Parameters: alpha : float
Constant that multiplies the penalty terms. Defaults to 1.0 See the notes for the exact mathematical meaning of this parameter.
alpha = 0is equivalent to an ordinary least square, solved by theLinearRegressionobject. For numerical reasons, usingalpha = 0with the Lasso object is not advised and you should prefer the LinearRegression object.l1_ratio : float
The ElasticNet mixing parameter, with
0 <= l1_ratio <= 1. Forl1_ratio = 0the penalty is an L2 penalty.For l1_ratio = 1it is an L1 penalty. For0 < l1_ratio < 1, the penalty is a combination of L1 and L2.fit_intercept : bool
Whether the intercept should be estimated or not. If
False, the data is assumed to be already centered.normalize : boolean, optional, default False
If
True, the regressors X will be normalized before regression.precompute : True | False | ‘auto’ | array-like
Whether to use a precomputed Gram matrix to speed up calculations. If set to
'auto'let us decide. The Gram matrix can also be passed as argument. For sparse input this option is alwaysTrueto preserve sparsity. WARNING : The'auto'option is deprecated and will be removed in 0.18.max_iter : int, optional
The maximum number of iterations
copy_X : boolean, optional, default True
If
True, X will be copied; else, it may be overwritten.tol : float, optional
The tolerance for the optimization: if the updates are smaller than
tol, the optimization code checks the dual gap for optimality and continues until it is smaller thantol.warm_start : bool, optional
When set to
True, reuse the solution of the previous call to fit as initialization, otherwise, just erase the previous solution.positive : bool, optional
When set to
True, forces the coefficients to be positive.selection : str, default ‘cyclic’
If set to ‘random’, a random coefficient is updated every iteration rather than looping over features sequentially by default. This (setting to ‘random’) often leads to significantly faster convergence especially when tol is higher than 1e-4.
random_state : int, RandomState instance, or None (default)
The seed of the pseudo random number generator that selects a random feature to update. Useful only when selection is set to ‘random’.
Attributes: coef_ : array, shape (n_features,) | (n_targets, n_features)
parameter vector (w in the cost function formula)
sparse_coef_ : scipy.sparse matrix, shape (n_features, 1) | (n_targets, n_features)
sparse_coef_is a readonly property derived fromcoef_intercept_ : float | array, shape (n_targets,)
independent term in decision function.
n_iter_ : array-like, shape (n_targets,)
number of iterations run by the coordinate descent solver to reach the specified tolerance.
See also
SGDRegressor- implements elastic net regression with incremental training.
SGDClassifier- implements logistic regression with elastic net penalty (
SGDClassifier(loss="log", penalty="elasticnet")).
Notes
To avoid unnecessary memory duplication the X argument of the fit method should be directly passed as a Fortran-contiguous numpy array.
Methods
decision_function(*args, **kwargs)DEPRECATED: and will be removed in 0.19 fit(X, y[, check_input])Fit model with coordinate descent. get_params([deep])Get parameters for this estimator. path(X, y[, l1_ratio, eps, n_alphas, ...])Compute elastic net path with coordinate descent predict(X)Predict using the linear model score(X, y[, sample_weight])Returns the coefficient of determination R^2 of the prediction. set_params(**params)Set the parameters of this estimator. -
__init__(alpha=1.0, l1_ratio=0.5, fit_intercept=True, normalize=False, precompute=False, max_iter=1000, copy_X=True, tol=0.0001, warm_start=False, positive=False, random_state=None, selection='cyclic')[source]¶
-
decision_function(*args, **kwargs)[source]¶ DEPRECATED: and will be removed in 0.19
Decision function of the linear model
Parameters: X : numpy array or scipy.sparse matrix of shape (n_samples, n_features)
Returns: T : array, shape (n_samples,)
The predicted decision function
-
fit(X, y, check_input=True)[source]¶ Fit model with coordinate descent.
Parameters: X : ndarray or scipy.sparse matrix, (n_samples, n_features)
Data
y : ndarray, shape (n_samples,) or (n_samples, n_targets)
Target
Notes
Coordinate descent is an algorithm that considers each column of data at a time hence it will automatically convert the X input as a Fortran-contiguous numpy array if necessary.
To avoid memory re-allocation it is advised to allocate the initial data in memory directly using that format.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
-
static
path(X, y, l1_ratio=0.5, eps=0.001, n_alphas=100, alphas=None, precompute='auto', Xy=None, copy_X=True, coef_init=None, verbose=False, return_n_iter=False, positive=False, check_input=True, **params)[source]¶ Compute elastic net path with coordinate descent
The elastic net optimization function varies for mono and multi-outputs.
For mono-output tasks it is:
1 / (2 * n_samples) * ||y - Xw||^2_2 + + alpha * l1_ratio * ||w||_1 + 0.5 * alpha * (1 - l1_ratio) * ||w||^2_2
For multi-output tasks it is:
(1 / (2 * n_samples)) * ||Y - XW||^Fro_2 + alpha * l1_ratio * ||W||_21 + 0.5 * alpha * (1 - l1_ratio) * ||W||_Fro^2
Where:
||W||_21 = \sum_i \sqrt{\sum_j w_{ij}^2}i.e. the sum of norm of each row.
Read more in the User Guide.
Parameters: X : {array-like}, shape (n_samples, n_features)
Training data. Pass directly as Fortran-contiguous data to avoid unnecessary memory duplication. If
yis mono-output thenXcan be sparse.y : ndarray, shape (n_samples,) or (n_samples, n_outputs)
Target values
l1_ratio : float, optional
float between 0 and 1 passed to elastic net (scaling between l1 and l2 penalties).
l1_ratio=1corresponds to the Lassoeps : float
Length of the path.
eps=1e-3means thatalpha_min / alpha_max = 1e-3n_alphas : int, optional
Number of alphas along the regularization path
alphas : ndarray, optional
List of alphas where to compute the models. If None alphas are set automatically
precompute : True | False | ‘auto’ | array-like
Whether to use a precomputed Gram matrix to speed up calculations. If set to
'auto'let us decide. The Gram matrix can also be passed as argument.Xy : array-like, optional
Xy = np.dot(X.T, y) that can be precomputed. It is useful only when the Gram matrix is precomputed.
copy_X : boolean, optional, default True
If
True, X will be copied; else, it may be overwritten.coef_init : array, shape (n_features, ) | None
The initial values of the coefficients.
verbose : bool or integer
Amount of verbosity.
params : kwargs
keyword arguments passed to the coordinate descent solver.
return_n_iter : bool
whether to return the number of iterations or not.
positive : bool, default False
If set to True, forces coefficients to be positive.
check_input : bool, default True
Skip input validation checks, including the Gram matrix when provided assuming there are handled by the caller when check_input=False.
Returns: alphas : array, shape (n_alphas,)
The alphas along the path where models are computed.
coefs : array, shape (n_features, n_alphas) or (n_outputs, n_features, n_alphas)
Coefficients along the path.
dual_gaps : array, shape (n_alphas,)
The dual gaps at the end of the optimization for each alpha.
n_iters : array-like, shape (n_alphas,)
The number of iterations taken by the coordinate descent optimizer to reach the specified tolerance for each alpha. (Is returned when
return_n_iteris set to True).Notes
See examples/plot_lasso_coordinate_descent_path.py for an example.
-
predict(X)[source]¶ Predict using the linear model
Parameters: X : {array-like, sparse matrix}, shape = (n_samples, n_features)
Samples.
Returns: C : array, shape = (n_samples,)
Returns predicted values.
-
score(X, y, sample_weight=None)[source]¶ Returns the coefficient of determination R^2 of the prediction.
The coefficient R^2 is defined as (1 - u/v), where u is the regression sum of squares ((y_true - y_pred) ** 2).sum() and v is the residual sum of squares ((y_true - y_true.mean()) ** 2).sum(). Best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value of y, disregarding the input features, would get a R^2 score of 0.0.
Parameters: X : array-like, shape = (n_samples, n_features)
Test samples.
y : array-like, shape = (n_samples) or (n_samples, n_outputs)
True values for X.
sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns: score : float
R^2 of self.predict(X) wrt. y.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The former have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: self :
-
sparse_coef_¶ sparse representation of the fitted coef