sklearn.mixture.GMM¶
- class sklearn.mixture.GMM(n_components=1, covariance_type='diag', random_state=None, thresh=0.01, min_covar=0.001, n_iter=100, n_init=1, params='wmc', init_params='wmc')¶
Gaussian Mixture Model
Representation of a Gaussian mixture model probability distribution. This class allows for easy evaluation of, sampling from, and maximum-likelihood estimation of the parameters of a GMM distribution.
Initializes parameters such that every mixture component has zero mean and identity covariance.
Parameters: n_components : int, optional
Number of mixture components. Defaults to 1.
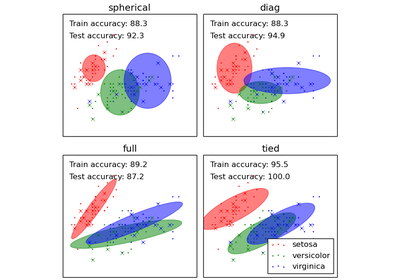
covariance_type : string, optional
String describing the type of covariance parameters to use. Must be one of ‘spherical’, ‘tied’, ‘diag’, ‘full’. Defaults to ‘diag’.
random_state: RandomState or an int seed (0 by default) :
A random number generator instance
min_covar : float, optional
Floor on the diagonal of the covariance matrix to prevent overfitting. Defaults to 1e-3.
thresh : float, optional
Convergence threshold.
n_iter : int, optional
Number of EM iterations to perform.
n_init : int, optional
Number of initializations to perform. the best results is kept
params : string, optional
Controls which parameters are updated in the training process. Can contain any combination of ‘w’ for weights, ‘m’ for means, and ‘c’ for covars. Defaults to ‘wmc’.
init_params : string, optional
Controls which parameters are updated in the initialization process. Can contain any combination of ‘w’ for weights, ‘m’ for means, and ‘c’ for covars. Defaults to ‘wmc’.
Attributes: `weights_` : array, shape (n_components,)
This attribute stores the mixing weights for each mixture component.
`means_` : array, shape (n_components, n_features)
Mean parameters for each mixture component.
`covars_` : array
Covariance parameters for each mixture component. The shape depends on covariance_type:
(n_components, n_features) if 'spherical', (n_features, n_features) if 'tied', (n_components, n_features) if 'diag', (n_components, n_features, n_features) if 'full'
`converged_` : bool
True when convergence was reached in fit(), False otherwise.
See also
Examples
>>> import numpy as np >>> from sklearn import mixture >>> np.random.seed(1) >>> g = mixture.GMM(n_components=2) >>> # Generate random observations with two modes centered on 0 >>> # and 10 to use for training. >>> obs = np.concatenate((np.random.randn(100, 1), ... 10 + np.random.randn(300, 1))) >>> g.fit(obs) GMM(covariance_type='diag', init_params='wmc', min_covar=0.001, n_components=2, n_init=1, n_iter=100, params='wmc', random_state=None, thresh=0.01) >>> np.round(g.weights_, 2) array([ 0.75, 0.25]) >>> np.round(g.means_, 2) array([[ 10.05], [ 0.06]]) >>> np.round(g.covars_, 2) array([[[ 1.02]], [[ 0.96]]]) >>> g.predict([[0], [2], [9], [10]]) array([1, 1, 0, 0]...) >>> np.round(g.score([[0], [2], [9], [10]]), 2) array([-2.19, -4.58, -1.75, -1.21]) >>> # Refit the model on new data (initial parameters remain the >>> # same), this time with an even split between the two modes. >>> g.fit(20 * [[0]] + 20 * [[10]]) GMM(covariance_type='diag', init_params='wmc', min_covar=0.001, n_components=2, n_init=1, n_iter=100, params='wmc', random_state=None, thresh=0.01) >>> np.round(g.weights_, 2) array([ 0.5, 0.5])
Methods
aic(X) Akaike information criterion for the current model fit bic(X) Bayesian information criterion for the current model fit eval(*args, **kwargs) DEPRECATED: GMM.eval was renamed to GMM.score_samples in 0.14 and will be removed in 0.16. fit(X) Estimate model parameters with the expectation-maximization algorithm. get_params([deep]) Get parameters for this estimator. predict(X) Predict label for data. predict_proba(X) Predict posterior probability of data under each Gaussian in the model. sample([n_samples, random_state]) Generate random samples from the model. score(X) Compute the log probability under the model. score_samples(X) Return the per-sample likelihood of the data under the model. set_params(**params) Set the parameters of this estimator. - __init__(n_components=1, covariance_type='diag', random_state=None, thresh=0.01, min_covar=0.001, n_iter=100, n_init=1, params='wmc', init_params='wmc')¶
- aic(X)¶
Akaike information criterion for the current model fit and the proposed data
Parameters: X : array of shape(n_samples, n_dimensions) Returns: aic: float (the lower the better) :
- bic(X)¶
Bayesian information criterion for the current model fit and the proposed data
Parameters: X : array of shape(n_samples, n_dimensions) Returns: bic: float (the lower the better) :
- eval(*args, **kwargs)¶
DEPRECATED: GMM.eval was renamed to GMM.score_samples in 0.14 and will be removed in 0.16.
- fit(X)¶
Estimate model parameters with the expectation-maximization algorithm.
A initialization step is performed before entering the em algorithm. If you want to avoid this step, set the keyword argument init_params to the empty string ‘’ when creating the GMM object. Likewise, if you would like just to do an initialization, set n_iter=0.
Parameters: X : array_like, shape (n, n_features)
List of n_features-dimensional data points. Each row corresponds to a single data point.
- get_params(deep=True)¶
Get parameters for this estimator.
Parameters: deep: boolean, optional :
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: params : mapping of string to any
Parameter names mapped to their values.
- predict(X)¶
Predict label for data.
Parameters: X : array-like, shape = [n_samples, n_features] Returns: C : array, shape = (n_samples,)
- predict_proba(X)¶
Predict posterior probability of data under each Gaussian in the model.
Parameters: X : array-like, shape = [n_samples, n_features]
Returns: responsibilities : array-like, shape = (n_samples, n_components)
Returns the probability of the sample for each Gaussian (state) in the model.
- sample(n_samples=1, random_state=None)¶
Generate random samples from the model.
Parameters: n_samples : int, optional
Number of samples to generate. Defaults to 1.
Returns: X : array_like, shape (n_samples, n_features)
List of samples
- score(X)¶
Compute the log probability under the model.
Parameters: X : array_like, shape (n_samples, n_features)
List of n_features-dimensional data points. Each row corresponds to a single data point.
Returns: logprob : array_like, shape (n_samples,)
Log probabilities of each data point in X
- score_samples(X)¶
Return the per-sample likelihood of the data under the model.
Compute the log probability of X under the model and return the posterior distribution (responsibilities) of each mixture component for each element of X.
Parameters: X: array_like, shape (n_samples, n_features) :
List of n_features-dimensional data points. Each row corresponds to a single data point.
Returns: logprob : array_like, shape (n_samples,)
Log probabilities of each data point in X.
responsibilities : array_like, shape (n_samples, n_components)
Posterior probabilities of each mixture component for each observation
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The former have parameters of the form <component>__<parameter> so that it’s possible to update each component of a nested object.
Returns: self :