sklearn.model_selection.StratifiedShuffleSplit¶
- class sklearn.model_selection.StratifiedShuffleSplit(n_splits=10, *, test_size=None, train_size=None, random_state=None)[source]¶
Stratified ShuffleSplit cross-validator
Provides train/test indices to split data in train/test sets.
This cross-validation object is a merge of StratifiedKFold and ShuffleSplit, which returns stratified randomized folds. The folds are made by preserving the percentage of samples for each class.
Note: like the ShuffleSplit strategy, stratified random splits do not guarantee that all folds will be different, although this is still very likely for sizeable datasets.
Read more in the User Guide.
- Parameters:
- n_splitsint, default=10
Number of re-shuffling & splitting iterations.
- test_sizefloat or int, default=None
If float, should be between 0.0 and 1.0 and represent the proportion of the dataset to include in the test split. If int, represents the absolute number of test samples. If None, the value is set to the complement of the train size. If
train_sizeis also None, it will be set to 0.1.- train_sizefloat or int, default=None
If float, should be between 0.0 and 1.0 and represent the proportion of the dataset to include in the train split. If int, represents the absolute number of train samples. If None, the value is automatically set to the complement of the test size.
- random_stateint, RandomState instance or None, default=None
Controls the randomness of the training and testing indices produced. Pass an int for reproducible output across multiple function calls. See Glossary.
Examples
>>> import numpy as np >>> from sklearn.model_selection import StratifiedShuffleSplit >>> X = np.array([[1, 2], [3, 4], [1, 2], [3, 4], [1, 2], [3, 4]]) >>> y = np.array([0, 0, 0, 1, 1, 1]) >>> sss = StratifiedShuffleSplit(n_splits=5, test_size=0.5, random_state=0) >>> sss.get_n_splits(X, y) 5 >>> print(sss) StratifiedShuffleSplit(n_splits=5, random_state=0, ...) >>> for i, (train_index, test_index) in enumerate(sss.split(X, y)): ... print(f"Fold {i}:") ... print(f" Train: index={train_index}") ... print(f" Test: index={test_index}") Fold 0: Train: index=[5 2 3] Test: index=[4 1 0] Fold 1: Train: index=[5 1 4] Test: index=[0 2 3] Fold 2: Train: index=[5 0 2] Test: index=[4 3 1] Fold 3: Train: index=[4 1 0] Test: index=[2 3 5] Fold 4: Train: index=[0 5 1] Test: index=[3 4 2]
Methods
get_n_splits([X, y, groups])Returns the number of splitting iterations in the cross-validator
split(X, y[, groups])Generate indices to split data into training and test set.
- get_n_splits(X=None, y=None, groups=None)[source]¶
Returns the number of splitting iterations in the cross-validator
- Parameters:
- Xobject
Always ignored, exists for compatibility.
- yobject
Always ignored, exists for compatibility.
- groupsobject
Always ignored, exists for compatibility.
- Returns:
- n_splitsint
Returns the number of splitting iterations in the cross-validator.
- split(X, y, groups=None)[source]¶
Generate indices to split data into training and test set.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Training data, where
n_samplesis the number of samples andn_featuresis the number of features.Note that providing
yis sufficient to generate the splits and hencenp.zeros(n_samples)may be used as a placeholder forXinstead of actual training data.- yarray-like of shape (n_samples,) or (n_samples, n_labels)
The target variable for supervised learning problems. Stratification is done based on the y labels.
- groupsobject
Always ignored, exists for compatibility.
- Yields:
- trainndarray
The training set indices for that split.
- testndarray
The testing set indices for that split.
Notes
Randomized CV splitters may return different results for each call of split. You can make the results identical by setting
random_stateto an integer.
Examples using sklearn.model_selection.StratifiedShuffleSplit¶
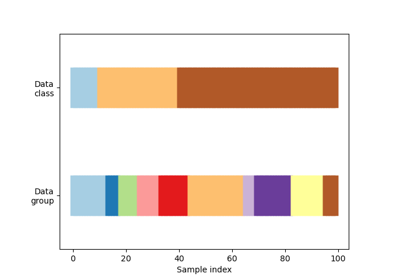
Visualizing cross-validation behavior in scikit-learn