sklearn.cluster.SpectralCoclustering¶
- class sklearn.cluster.SpectralCoclustering(n_clusters=3, *, svd_method='randomized', n_svd_vecs=None, mini_batch=False, init='k-means++', n_init=10, random_state=None)[source]¶
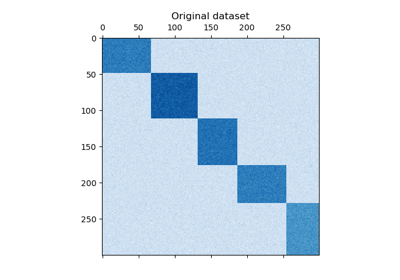
Spectral Co-Clustering algorithm (Dhillon, 2001).
Clusters rows and columns of an array
Xto solve the relaxed normalized cut of the bipartite graph created fromXas follows: the edge between row vertexiand column vertexjhas weightX[i, j].The resulting bicluster structure is block-diagonal, since each row and each column belongs to exactly one bicluster.
Supports sparse matrices, as long as they are nonnegative.
Read more in the User Guide.
- Parameters:
- n_clustersint, default=3
The number of biclusters to find.
- svd_method{‘randomized’, ‘arpack’}, default=’randomized’
Selects the algorithm for finding singular vectors. May be ‘randomized’ or ‘arpack’. If ‘randomized’, use
sklearn.utils.extmath.randomized_svd, which may be faster for large matrices. If ‘arpack’, usescipy.sparse.linalg.svds, which is more accurate, but possibly slower in some cases.- n_svd_vecsint, default=None
Number of vectors to use in calculating the SVD. Corresponds to
ncvwhensvd_method=arpackandn_oversampleswhensvd_methodis ‘randomized`.- mini_batchbool, default=False
Whether to use mini-batch k-means, which is faster but may get different results.
- init{‘k-means++’, ‘random’}, or ndarray of shape (n_clusters, n_features), default=’k-means++’
Method for initialization of k-means algorithm; defaults to ‘k-means++’.
- n_initint, default=10
Number of random initializations that are tried with the k-means algorithm.
If mini-batch k-means is used, the best initialization is chosen and the algorithm runs once. Otherwise, the algorithm is run for each initialization and the best solution chosen.
- random_stateint, RandomState instance, default=None
Used for randomizing the singular value decomposition and the k-means initialization. Use an int to make the randomness deterministic. See Glossary.
- Attributes:
- rows_array-like of shape (n_row_clusters, n_rows)
Results of the clustering.
rows[i, r]is True if clustericontains rowr. Available only after callingfit.- columns_array-like of shape (n_column_clusters, n_columns)
Results of the clustering, like
rows.- row_labels_array-like of shape (n_rows,)
The bicluster label of each row.
- column_labels_array-like of shape (n_cols,)
The bicluster label of each column.
biclusters_tuple of two ndarraysConvenient way to get row and column indicators together.
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
SpectralBiclusteringPartitions rows and columns under the assumption that the data has an underlying checkerboard structure.
References
Examples
>>> from sklearn.cluster import SpectralCoclustering >>> import numpy as np >>> X = np.array([[1, 1], [2, 1], [1, 0], ... [4, 7], [3, 5], [3, 6]]) >>> clustering = SpectralCoclustering(n_clusters=2, random_state=0).fit(X) >>> clustering.row_labels_ array([0, 1, 1, 0, 0, 0], dtype=int32) >>> clustering.column_labels_ array([0, 0], dtype=int32) >>> clustering SpectralCoclustering(n_clusters=2, random_state=0)
Methods
fit(X[, y])Create a biclustering for X.
get_indices(i)Row and column indices of the
i'th bicluster.get_params([deep])Get parameters for this estimator.
get_shape(i)Shape of the
i'th bicluster.get_submatrix(i, data)Return the submatrix corresponding to bicluster
i.set_params(**params)Set the parameters of this estimator.
- property biclusters_¶
Convenient way to get row and column indicators together.
Returns the
rows_andcolumns_members.
- fit(X, y=None)[source]¶
Create a biclustering for X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Training data.
- yIgnored
Not used, present for API consistency by convention.
- Returns:
- selfobject
SpectralBiclustering instance.
- get_indices(i)[source]¶
Row and column indices of the
i’th bicluster.Only works if
rows_andcolumns_attributes exist.- Parameters:
- iint
The index of the cluster.
- Returns:
- row_indndarray, dtype=np.intp
Indices of rows in the dataset that belong to the bicluster.
- col_indndarray, dtype=np.intp
Indices of columns in the dataset that belong to the bicluster.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- get_shape(i)[source]¶
Shape of the
i’th bicluster.- Parameters:
- iint
The index of the cluster.
- Returns:
- n_rowsint
Number of rows in the bicluster.
- n_colsint
Number of columns in the bicluster.
- get_submatrix(i, data)[source]¶
Return the submatrix corresponding to bicluster
i.- Parameters:
- iint
The index of the cluster.
- dataarray-like of shape (n_samples, n_features)
The data.
- Returns:
- submatrixndarray of shape (n_rows, n_cols)
The submatrix corresponding to bicluster
i.
Notes
Works with sparse matrices. Only works if
rows_andcolumns_attributes exist.
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using sklearn.cluster.SpectralCoclustering¶
Biclustering documents with the Spectral Co-clustering algorithm