Note
Click here to download the full example code or to run this example in your browser via Binder
Sparsity Example: Fitting only features 1 and 2¶
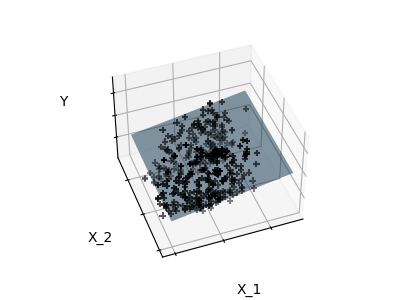
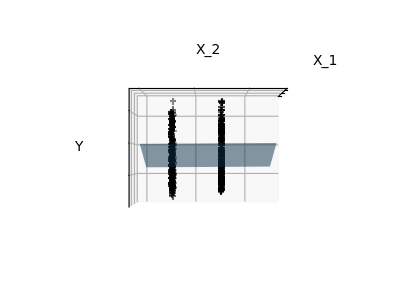
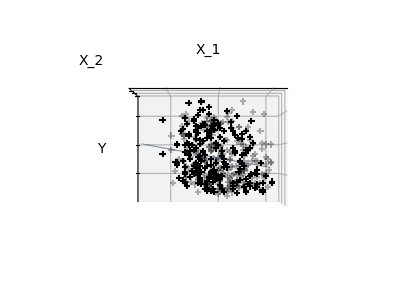
Features 1 and 2 of the diabetes-dataset are fitted and
plotted below. It illustrates that although feature 2
has a strong coefficient on the full model, it does not
give us much regarding y when compared to just feature 1.
# Code source: Gaël Varoquaux
# Modified for documentation by Jaques Grobler
# License: BSD 3 clause
First we load the diabetes dataset.
from sklearn import datasets
import numpy as np
X, y = datasets.load_diabetes(return_X_y=True)
indices = (0, 1)
X_train = X[:-20, indices]
X_test = X[-20:, indices]
y_train = y[:-20]
y_test = y[-20:]
Next we fit a linear regression model.
from sklearn import linear_model
ols = linear_model.LinearRegression()
_ = ols.fit(X_train, y_train)
Finally we plot the figure from three different views.
import matplotlib.pyplot as plt
# unused but required import for doing 3d projections with matplotlib < 3.2
import mpl_toolkits.mplot3d # noqa: F401
def plot_figs(fig_num, elev, azim, X_train, clf):
fig = plt.figure(fig_num, figsize=(4, 3))
plt.clf()
ax = fig.add_subplot(111, projection="3d", elev=elev, azim=azim)
ax.scatter(X_train[:, 0], X_train[:, 1], y_train, c="k", marker="+")
ax.plot_surface(
np.array([[-0.1, -0.1], [0.15, 0.15]]),
np.array([[-0.1, 0.15], [-0.1, 0.15]]),
clf.predict(
np.array([[-0.1, -0.1, 0.15, 0.15], [-0.1, 0.15, -0.1, 0.15]]).T
).reshape((2, 2)),
alpha=0.5,
)
ax.set_xlabel("X_1")
ax.set_ylabel("X_2")
ax.set_zlabel("Y")
ax.xaxis.set_ticklabels([])
ax.yaxis.set_ticklabels([])
ax.zaxis.set_ticklabels([])
# Generate the three different figures from different views
elev = 43.5
azim = -110
plot_figs(1, elev, azim, X_train, ols)
elev = -0.5
azim = 0
plot_figs(2, elev, azim, X_train, ols)
elev = -0.5
azim = 90
plot_figs(3, elev, azim, X_train, ols)
plt.show()
Total running time of the script: ( 0 minutes 0.251 seconds)