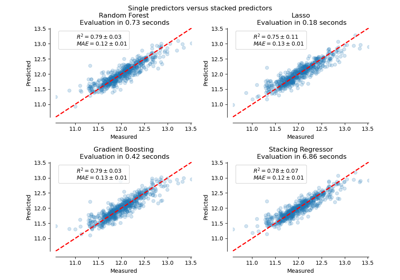
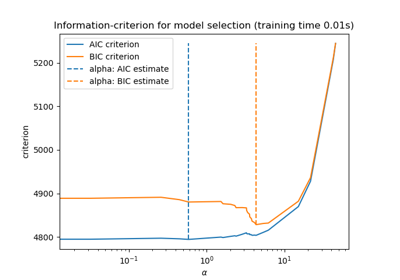
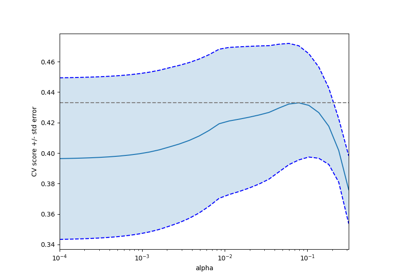
sklearn.linear_model.LassoCV¶
- class sklearn.linear_model.LassoCV(*, eps=0.001, n_alphas=100, alphas=None, fit_intercept=True, normalize='deprecated', precompute='auto', max_iter=1000, tol=0.0001, copy_X=True, cv=None, verbose=False, n_jobs=None, positive=False, random_state=None, selection='cyclic')[source]¶
Lasso linear model with iterative fitting along a regularization path.
See glossary entry for cross-validation estimator.
The best model is selected by cross-validation.
The optimization objective for Lasso is:
(1 / (2 * n_samples)) * ||y - Xw||^2_2 + alpha * ||w||_1
Read more in the User Guide.
- Parameters:
- epsfloat, default=1e-3
Length of the path.
eps=1e-3means thatalpha_min / alpha_max = 1e-3.- n_alphasint, default=100
Number of alphas along the regularization path.
- alphasndarray, default=None
List of alphas where to compute the models. If
Nonealphas are set automatically.- fit_interceptbool, default=True
Whether to calculate the intercept for this model. If set to false, no intercept will be used in calculations (i.e. data is expected to be centered).
- normalizebool, default=False
This parameter is ignored when
fit_interceptis set to False. If True, the regressors X will be normalized before regression by subtracting the mean and dividing by the l2-norm. If you wish to standardize, please useStandardScalerbefore callingfiton an estimator withnormalize=False.Deprecated since version 1.0:
normalizewas deprecated in version 1.0 and will be removed in 1.2.- precompute‘auto’, bool or array-like of shape (n_features, n_features), default=’auto’
Whether to use a precomputed Gram matrix to speed up calculations. If set to
'auto'let us decide. The Gram matrix can also be passed as argument.- max_iterint, default=1000
The maximum number of iterations.
- tolfloat, default=1e-4
The tolerance for the optimization: if the updates are smaller than
tol, the optimization code checks the dual gap for optimality and continues until it is smaller thantol.- copy_Xbool, default=True
If
True, X will be copied; else, it may be overwritten.- cvint, cross-validation generator or iterable, default=None
Determines the cross-validation splitting strategy. Possible inputs for cv are:
None, to use the default 5-fold cross-validation,
int, to specify the number of folds.
An iterable yielding (train, test) splits as arrays of indices.
For int/None inputs,
KFoldis used.Refer User Guide for the various cross-validation strategies that can be used here.
Changed in version 0.22:
cvdefault value if None changed from 3-fold to 5-fold.- verbosebool or int, default=False
Amount of verbosity.
- n_jobsint, default=None
Number of CPUs to use during the cross validation.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- positivebool, default=False
If positive, restrict regression coefficients to be positive.
- random_stateint, RandomState instance, default=None
The seed of the pseudo random number generator that selects a random feature to update. Used when
selection== ‘random’. Pass an int for reproducible output across multiple function calls. See Glossary.- selection{‘cyclic’, ‘random’}, default=’cyclic’
If set to ‘random’, a random coefficient is updated every iteration rather than looping over features sequentially by default. This (setting to ‘random’) often leads to significantly faster convergence especially when tol is higher than 1e-4.
- Attributes:
- alpha_float
The amount of penalization chosen by cross validation.
- coef_ndarray of shape (n_features,) or (n_targets, n_features)
Parameter vector (w in the cost function formula).
- intercept_float or ndarray of shape (n_targets,)
Independent term in decision function.
- mse_path_ndarray of shape (n_alphas, n_folds)
Mean square error for the test set on each fold, varying alpha.
- alphas_ndarray of shape (n_alphas,)
The grid of alphas used for fitting.
- dual_gap_float or ndarray of shape (n_targets,)
The dual gap at the end of the optimization for the optimal alpha (
alpha_).- n_iter_int
Number of iterations run by the coordinate descent solver to reach the specified tolerance for the optimal alpha.
- n_features_in_int
Number of features seen during fit.
New in version 0.24.
- feature_names_in_ndarray of shape (
n_features_in_,) Names of features seen during fit. Defined only when
Xhas feature names that are all strings.New in version 1.0.
See also
lars_pathCompute Least Angle Regression or Lasso path using LARS algorithm.
lasso_pathCompute Lasso path with coordinate descent.
LassoThe Lasso is a linear model that estimates sparse coefficients.
LassoLarsLasso model fit with Least Angle Regression a.k.a. Lars.
LassoCVLasso linear model with iterative fitting along a regularization path.
LassoLarsCVCross-validated Lasso using the LARS algorithm.
Notes
In
fit, once the best parameteralphais found through cross-validation, the model is fit again using the entire training set.To avoid unnecessary memory duplication the
Xargument of thefitmethod should be directly passed as a Fortran-contiguous numpy array.For an example, see examples/linear_model/plot_lasso_model_selection.py.
Examples
>>> from sklearn.linear_model import LassoCV >>> from sklearn.datasets import make_regression >>> X, y = make_regression(noise=4, random_state=0) >>> reg = LassoCV(cv=5, random_state=0).fit(X, y) >>> reg.score(X, y) 0.9993... >>> reg.predict(X[:1,]) array([-78.4951...])
Methods
fit(X, y[, sample_weight])Fit linear model with coordinate descent.
get_params([deep])Get parameters for this estimator.
path(X, y, *[, eps, n_alphas, alphas, ...])Compute Lasso path with coordinate descent.
predict(X)Predict using the linear model.
score(X, y[, sample_weight])Return the coefficient of determination of the prediction.
set_params(**params)Set the parameters of this estimator.
- fit(X, y, sample_weight=None)[source]¶
Fit linear model with coordinate descent.
Fit is on grid of alphas and best alpha estimated by cross-validation.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Training data. Pass directly as Fortran-contiguous data to avoid unnecessary memory duplication. If y is mono-output, X can be sparse.
- yarray-like of shape (n_samples,) or (n_samples, n_targets)
Target values.
- sample_weightfloat or array-like of shape (n_samples,), default=None
Sample weights used for fitting and evaluation of the weighted mean squared error of each cv-fold. Note that the cross validated MSE that is finally used to find the best model is the unweighted mean over the (weighted) MSEs of each test fold.
- Returns:
- selfobject
Returns an instance of fitted model.
- get_params(deep=True)[source]¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- static path(X, y, *, eps=0.001, n_alphas=100, alphas=None, precompute='auto', Xy=None, copy_X=True, coef_init=None, verbose=False, return_n_iter=False, positive=False, **params)[source]¶
Compute Lasso path with coordinate descent.
The Lasso optimization function varies for mono and multi-outputs.
For mono-output tasks it is:
(1 / (2 * n_samples)) * ||y - Xw||^2_2 + alpha * ||w||_1
For multi-output tasks it is:
(1 / (2 * n_samples)) * ||Y - XW||^2_Fro + alpha * ||W||_21
Where:
||W||_21 = \sum_i \sqrt{\sum_j w_{ij}^2}
i.e. the sum of norm of each row.
Read more in the User Guide.
- Parameters:
- X{array-like, sparse matrix} of shape (n_samples, n_features)
Training data. Pass directly as Fortran-contiguous data to avoid unnecessary memory duplication. If
yis mono-output thenXcan be sparse.- y{array-like, sparse matrix} of shape (n_samples,) or (n_samples, n_targets)
Target values.
- epsfloat, default=1e-3
Length of the path.
eps=1e-3means thatalpha_min / alpha_max = 1e-3.- n_alphasint, default=100
Number of alphas along the regularization path.
- alphasndarray, default=None
List of alphas where to compute the models. If
Nonealphas are set automatically.- precompute‘auto’, bool or array-like of shape (n_features, n_features), default=’auto’
Whether to use a precomputed Gram matrix to speed up calculations. If set to
'auto'let us decide. The Gram matrix can also be passed as argument.- Xyarray-like of shape (n_features,) or (n_features, n_targets), default=None
Xy = np.dot(X.T, y) that can be precomputed. It is useful only when the Gram matrix is precomputed.
- copy_Xbool, default=True
If
True, X will be copied; else, it may be overwritten.- coef_initndarray of shape (n_features, ), default=None
The initial values of the coefficients.
- verbosebool or int, default=False
Amount of verbosity.
- return_n_iterbool, default=False
Whether to return the number of iterations or not.
- positivebool, default=False
If set to True, forces coefficients to be positive. (Only allowed when
y.ndim == 1).- **paramskwargs
Keyword arguments passed to the coordinate descent solver.
- Returns:
- alphasndarray of shape (n_alphas,)
The alphas along the path where models are computed.
- coefsndarray of shape (n_features, n_alphas) or (n_targets, n_features, n_alphas)
Coefficients along the path.
- dual_gapsndarray of shape (n_alphas,)
The dual gaps at the end of the optimization for each alpha.
- n_iterslist of int
The number of iterations taken by the coordinate descent optimizer to reach the specified tolerance for each alpha.
See also
lars_pathCompute Least Angle Regression or Lasso path using LARS algorithm.
LassoThe Lasso is a linear model that estimates sparse coefficients.
LassoLarsLasso model fit with Least Angle Regression a.k.a. Lars.
LassoCVLasso linear model with iterative fitting along a regularization path.
LassoLarsCVCross-validated Lasso using the LARS algorithm.
sklearn.decomposition.sparse_encodeEstimator that can be used to transform signals into sparse linear combination of atoms from a fixed.
Notes
For an example, see examples/linear_model/plot_lasso_coordinate_descent_path.py.
To avoid unnecessary memory duplication the X argument of the fit method should be directly passed as a Fortran-contiguous numpy array.
Note that in certain cases, the Lars solver may be significantly faster to implement this functionality. In particular, linear interpolation can be used to retrieve model coefficients between the values output by lars_path
Examples
Comparing lasso_path and lars_path with interpolation:
>>> import numpy as np >>> from sklearn.linear_model import lasso_path >>> X = np.array([[1, 2, 3.1], [2.3, 5.4, 4.3]]).T >>> y = np.array([1, 2, 3.1]) >>> # Use lasso_path to compute a coefficient path >>> _, coef_path, _ = lasso_path(X, y, alphas=[5., 1., .5]) >>> print(coef_path) [[0. 0. 0.46874778] [0.2159048 0.4425765 0.23689075]]
>>> # Now use lars_path and 1D linear interpolation to compute the >>> # same path >>> from sklearn.linear_model import lars_path >>> alphas, active, coef_path_lars = lars_path(X, y, method='lasso') >>> from scipy import interpolate >>> coef_path_continuous = interpolate.interp1d(alphas[::-1], ... coef_path_lars[:, ::-1]) >>> print(coef_path_continuous([5., 1., .5])) [[0. 0. 0.46915237] [0.2159048 0.4425765 0.23668876]]
- predict(X)[source]¶
Predict using the linear model.
- Parameters:
- Xarray-like or sparse matrix, shape (n_samples, n_features)
Samples.
- Returns:
- Carray, shape (n_samples,)
Returns predicted values.
- score(X, y, sample_weight=None)[source]¶
Return the coefficient of determination of the prediction.
The coefficient of determination \(R^2\) is defined as \((1 - \frac{u}{v})\), where \(u\) is the residual sum of squares
((y_true - y_pred)** 2).sum()and \(v\) is the total sum of squares((y_true - y_true.mean()) ** 2).sum(). The best possible score is 1.0 and it can be negative (because the model can be arbitrarily worse). A constant model that always predicts the expected value ofy, disregarding the input features, would get a \(R^2\) score of 0.0.- Parameters:
- Xarray-like of shape (n_samples, n_features)
Test samples. For some estimators this may be a precomputed kernel matrix or a list of generic objects instead with shape
(n_samples, n_samples_fitted), wheren_samples_fittedis the number of samples used in the fitting for the estimator.- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True values for
X.- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns:
- scorefloat
\(R^2\) of
self.predict(X)wrt.y.
Notes
The \(R^2\) score used when calling
scoreon a regressor usesmultioutput='uniform_average'from version 0.23 to keep consistent with default value ofr2_score. This influences thescoremethod of all the multioutput regressors (except forMultiOutputRegressor).
- set_params(**params)[source]¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using sklearn.linear_model.LassoCV¶
Common pitfalls in the interpretation of coefficients of linear models