Note
Click here to download the full example code or to run this example in your browser via Binder
Model-based and sequential feature selection¶
This example illustrates and compares two approaches for feature selection:
SelectFromModel which is based on feature
importance, and
SequentialFeatureSelection which relies
on a greedy approach.
We use the Diabetes dataset, which consists of 10 features collected from 442 diabetes patients.
Authors: Manoj Kumar, Maria Telenczuk, Nicolas Hug.
License: BSD 3 clause
Loading the data¶
We first load the diabetes dataset which is available from within scikit-learn, and print its description:
from sklearn.datasets import load_diabetes
diabetes = load_diabetes()
X, y = diabetes.data, diabetes.target
print(diabetes.DESCR)
.. _diabetes_dataset:
Diabetes dataset
----------------
Ten baseline variables, age, sex, body mass index, average blood
pressure, and six blood serum measurements were obtained for each of n =
442 diabetes patients, as well as the response of interest, a
quantitative measure of disease progression one year after baseline.
**Data Set Characteristics:**
:Number of Instances: 442
:Number of Attributes: First 10 columns are numeric predictive values
:Target: Column 11 is a quantitative measure of disease progression one year after baseline
:Attribute Information:
- age age in years
- sex
- bmi body mass index
- bp average blood pressure
- s1 tc, total serum cholesterol
- s2 ldl, low-density lipoproteins
- s3 hdl, high-density lipoproteins
- s4 tch, total cholesterol / HDL
- s5 ltg, possibly log of serum triglycerides level
- s6 glu, blood sugar level
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times the square root of `n_samples` (i.e. the sum of squares of each column totals 1).
Source URL:
https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html
For more information see:
Bradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) "Least Angle Regression," Annals of Statistics (with discussion), 407-499.
(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)
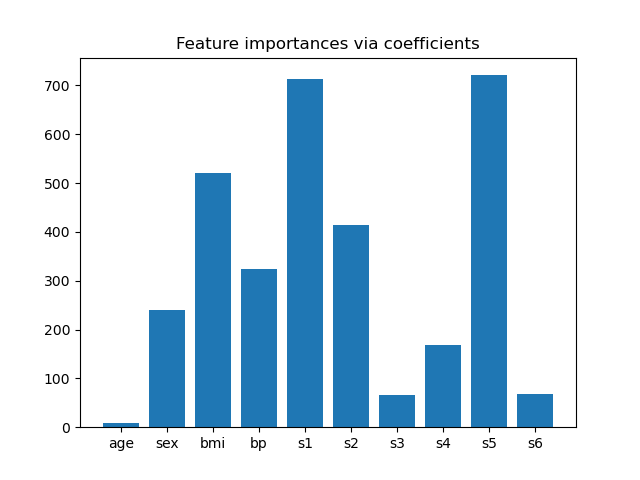
Feature importance from coefficients¶
To get an idea of the importance of the features, we are going to use the
RidgeCV estimator. The features with the
highest absolute coef_ value are considered the most important.
We can observe the coefficients directly without needing to scale them (or
scale the data) because from the description above, we know that the features
were already standardized.
For a more complete example on the interpretations of the coefficients of
linear models, you may refer to
Common pitfalls in the interpretation of coefficients of linear models.
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import RidgeCV
ridge = RidgeCV(alphas=np.logspace(-6, 6, num=5)).fit(X, y)
importance = np.abs(ridge.coef_)
feature_names = np.array(diabetes.feature_names)
plt.bar(height=importance, x=feature_names)
plt.title("Feature importances via coefficients")
plt.show()
Selecting features based on importance¶
Now we want to select the two features which are the most important according
to the coefficients. The SelectFromModel
is meant just for that. SelectFromModel
accepts a threshold parameter and will select the features whose importance
(defined by the coefficients) are above this threshold.
Since we want to select only 2 features, we will set this threshold slightly above the coefficient of third most important feature.
from sklearn.feature_selection import SelectFromModel
from time import time
threshold = np.sort(importance)[-3] + 0.01
tic = time()
sfm = SelectFromModel(ridge, threshold=threshold).fit(X, y)
toc = time()
print(f"Features selected by SelectFromModel: {feature_names[sfm.get_support()]}")
print(f"Done in {toc - tic:.3f}s")
Features selected by SelectFromModel: ['s1' 's5']
Done in 0.001s
Selecting features with Sequential Feature Selection¶
Another way of selecting features is to use
SequentialFeatureSelector
(SFS). SFS is a greedy procedure where, at each iteration, we choose the best
new feature to add to our selected features based a cross-validation score.
That is, we start with 0 features and choose the best single feature with the
highest score. The procedure is repeated until we reach the desired number of
selected features.
We can also go in the reverse direction (backward SFS), i.e. start with all the features and greedily choose features to remove one by one. We illustrate both approaches here.
from sklearn.feature_selection import SequentialFeatureSelector
tic_fwd = time()
sfs_forward = SequentialFeatureSelector(
ridge, n_features_to_select=2, direction="forward"
).fit(X, y)
toc_fwd = time()
tic_bwd = time()
sfs_backward = SequentialFeatureSelector(
ridge, n_features_to_select=2, direction="backward"
).fit(X, y)
toc_bwd = time()
print(
"Features selected by forward sequential selection: "
f"{feature_names[sfs_forward.get_support()]}"
)
print(f"Done in {toc_fwd - tic_fwd:.3f}s")
print(
"Features selected by backward sequential selection: "
f"{feature_names[sfs_backward.get_support()]}"
)
print(f"Done in {toc_bwd - tic_bwd:.3f}s")
Features selected by forward sequential selection: ['bmi' 's5']
Done in 0.114s
Features selected by backward sequential selection: ['bmi' 's5']
Done in 0.336s
Discussion¶
Interestingly, forward and backward selection have selected the same set of features. In general, this isn’t the case and the two methods would lead to different results.
We also note that the features selected by SFS differ from those selected by
feature importance: SFS selects bmi instead of s1. This does sound
reasonable though, since bmi corresponds to the third most important
feature according to the coefficients. It is quite remarkable considering
that SFS makes no use of the coefficients at all.
To finish with, we should note that
SelectFromModel is significantly faster
than SFS. Indeed, SelectFromModel only
needs to fit a model once, while SFS needs to cross-validate many different
models for each of the iterations. SFS however works with any model, while
SelectFromModel requires the underlying
estimator to expose a coef_ attribute or a feature_importances_
attribute. The forward SFS is faster than the backward SFS because it only
needs to perform n_features_to_select = 2 iterations, while the backward
SFS needs to perform n_features - n_features_to_select = 8 iterations.
Total running time of the script: ( 0 minutes 0.517 seconds)
