Note
Click here to download the full example code or to run this example in your browser via Binder
Comparison of the K-Means and MiniBatchKMeans clustering algorithms¶
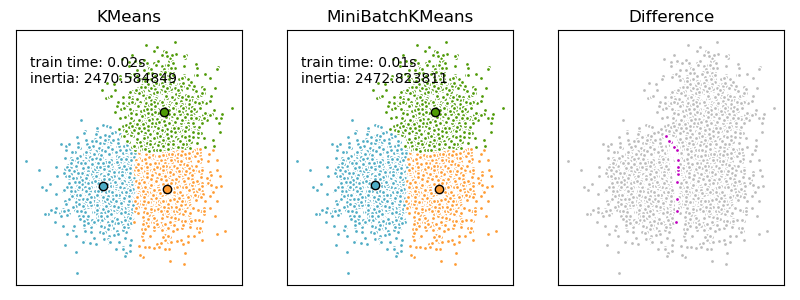
We want to compare the performance of the MiniBatchKMeans and KMeans: the MiniBatchKMeans is faster, but gives slightly different results (see Mini Batch K-Means).
We will cluster a set of data, first with KMeans and then with MiniBatchKMeans, and plot the results. We will also plot the points that are labelled differently between the two algorithms.
import time
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import MiniBatchKMeans, KMeans
from sklearn.metrics.pairwise import pairwise_distances_argmin
from sklearn.datasets import make_blobs
# #############################################################################
# Generate sample data
np.random.seed(0)
batch_size = 45
centers = [[1, 1], [-1, -1], [1, -1]]
n_clusters = len(centers)
X, labels_true = make_blobs(n_samples=3000, centers=centers, cluster_std=0.7)
# #############################################################################
# Compute clustering with Means
k_means = KMeans(init="k-means++", n_clusters=3, n_init=10)
t0 = time.time()
k_means.fit(X)
t_batch = time.time() - t0
# #############################################################################
# Compute clustering with MiniBatchKMeans
mbk = MiniBatchKMeans(
init="k-means++",
n_clusters=3,
batch_size=batch_size,
n_init=10,
max_no_improvement=10,
verbose=0,
)
t0 = time.time()
mbk.fit(X)
t_mini_batch = time.time() - t0
# #############################################################################
# Plot result
fig = plt.figure(figsize=(8, 3))
fig.subplots_adjust(left=0.02, right=0.98, bottom=0.05, top=0.9)
colors = ["#4EACC5", "#FF9C34", "#4E9A06"]
# We want to have the same colors for the same cluster from the
# MiniBatchKMeans and the KMeans algorithm. Let's pair the cluster centers per
# closest one.
k_means_cluster_centers = k_means.cluster_centers_
order = pairwise_distances_argmin(k_means.cluster_centers_, mbk.cluster_centers_)
mbk_means_cluster_centers = mbk.cluster_centers_[order]
k_means_labels = pairwise_distances_argmin(X, k_means_cluster_centers)
mbk_means_labels = pairwise_distances_argmin(X, mbk_means_cluster_centers)
# KMeans
ax = fig.add_subplot(1, 3, 1)
for k, col in zip(range(n_clusters), colors):
my_members = k_means_labels == k
cluster_center = k_means_cluster_centers[k]
ax.plot(X[my_members, 0], X[my_members, 1], "w", markerfacecolor=col, marker=".")
ax.plot(
cluster_center[0],
cluster_center[1],
"o",
markerfacecolor=col,
markeredgecolor="k",
markersize=6,
)
ax.set_title("KMeans")
ax.set_xticks(())
ax.set_yticks(())
plt.text(-3.5, 1.8, "train time: %.2fs\ninertia: %f" % (t_batch, k_means.inertia_))
# MiniBatchKMeans
ax = fig.add_subplot(1, 3, 2)
for k, col in zip(range(n_clusters), colors):
my_members = mbk_means_labels == k
cluster_center = mbk_means_cluster_centers[k]
ax.plot(X[my_members, 0], X[my_members, 1], "w", markerfacecolor=col, marker=".")
ax.plot(
cluster_center[0],
cluster_center[1],
"o",
markerfacecolor=col,
markeredgecolor="k",
markersize=6,
)
ax.set_title("MiniBatchKMeans")
ax.set_xticks(())
ax.set_yticks(())
plt.text(-3.5, 1.8, "train time: %.2fs\ninertia: %f" % (t_mini_batch, mbk.inertia_))
# Initialise the different array to all False
different = mbk_means_labels == 4
ax = fig.add_subplot(1, 3, 3)
for k in range(n_clusters):
different += (k_means_labels == k) != (mbk_means_labels == k)
identic = np.logical_not(different)
ax.plot(X[identic, 0], X[identic, 1], "w", markerfacecolor="#bbbbbb", marker=".")
ax.plot(X[different, 0], X[different, 1], "w", markerfacecolor="m", marker=".")
ax.set_title("Difference")
ax.set_xticks(())
ax.set_yticks(())
plt.show()
Total running time of the script: ( 0 minutes 0.185 seconds)
