Note
Click here to download the full example code or to run this example in your browser via Binder
K-means Clustering¶
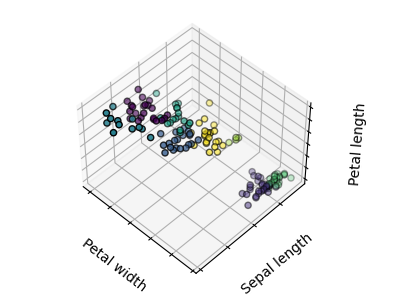
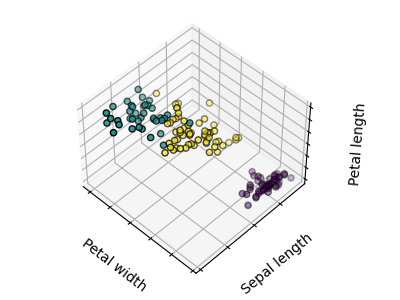
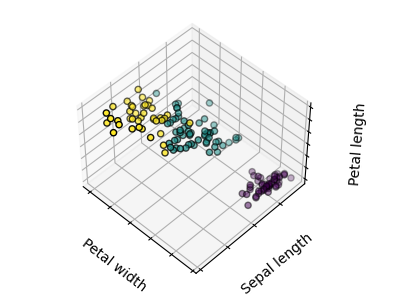
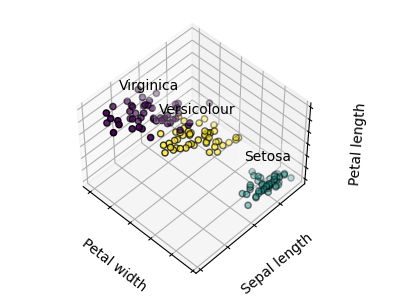
The plots display firstly what a K-means algorithm would yield using three clusters. It is then shown what the effect of a bad initialization is on the classification process: By setting n_init to only 1 (default is 10), the amount of times that the algorithm will be run with different centroid seeds is reduced. The next plot displays what using eight clusters would deliver and finally the ground truth.
Out:
/home/circleci/project/examples/cluster/plot_cluster_iris.py:48: MatplotlibDeprecationWarning: Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
/home/circleci/project/examples/cluster/plot_cluster_iris.py:48: MatplotlibDeprecationWarning: Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
/home/circleci/project/examples/cluster/plot_cluster_iris.py:48: MatplotlibDeprecationWarning: Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
/home/circleci/project/examples/cluster/plot_cluster_iris.py:66: MatplotlibDeprecationWarning: Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
# Code source: Gaël Varoquaux
# Modified for documentation by Jaques Grobler
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
# Though the following import is not directly being used, it is required
# for 3D projection to work
from mpl_toolkits.mplot3d import Axes3D
from sklearn.cluster import KMeans
from sklearn import datasets
np.random.seed(5)
iris = datasets.load_iris()
X = iris.data
y = iris.target
estimators = [
("k_means_iris_8", KMeans(n_clusters=8)),
("k_means_iris_3", KMeans(n_clusters=3)),
("k_means_iris_bad_init", KMeans(n_clusters=3, n_init=1, init="random")),
]
fignum = 1
titles = ["8 clusters", "3 clusters", "3 clusters, bad initialization"]
for name, est in estimators:
fig = plt.figure(fignum, figsize=(4, 3))
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
est.fit(X)
labels = est.labels_
ax.scatter(X[:, 3], X[:, 0], X[:, 2], c=labels.astype(float), edgecolor="k")
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel("Petal width")
ax.set_ylabel("Sepal length")
ax.set_zlabel("Petal length")
ax.set_title(titles[fignum - 1])
ax.dist = 12
fignum = fignum + 1
# Plot the ground truth
fig = plt.figure(fignum, figsize=(4, 3))
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=134)
for name, label in [("Setosa", 0), ("Versicolour", 1), ("Virginica", 2)]:
ax.text3D(
X[y == label, 3].mean(),
X[y == label, 0].mean(),
X[y == label, 2].mean() + 2,
name,
horizontalalignment="center",
bbox=dict(alpha=0.2, edgecolor="w", facecolor="w"),
)
# Reorder the labels to have colors matching the cluster results
y = np.choose(y, [1, 2, 0]).astype(float)
ax.scatter(X[:, 3], X[:, 0], X[:, 2], c=y, edgecolor="k")
ax.w_xaxis.set_ticklabels([])
ax.w_yaxis.set_ticklabels([])
ax.w_zaxis.set_ticklabels([])
ax.set_xlabel("Petal width")
ax.set_ylabel("Sepal length")
ax.set_zlabel("Petal length")
ax.set_title("Ground Truth")
ax.dist = 12
fig.show()
Total running time of the script: ( 0 minutes 0.310 seconds)