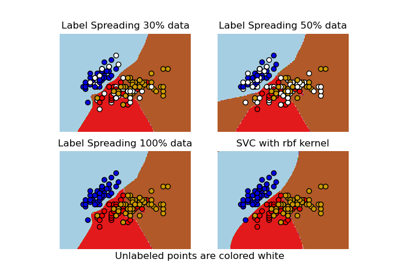
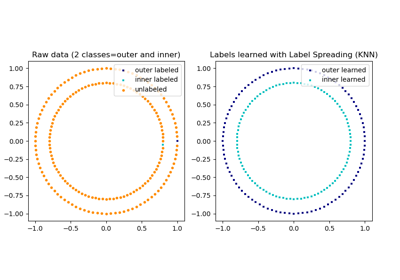
sklearn.semi_supervised.LabelSpreading¶
-
class
sklearn.semi_supervised.LabelSpreading(kernel='rbf', *, gamma=20, n_neighbors=7, alpha=0.2, max_iter=30, tol=0.001, n_jobs=None)[source]¶ LabelSpreading model for semi-supervised learning
This model is similar to the basic Label Propagation algorithm, but uses affinity matrix based on the normalized graph Laplacian and soft clamping across the labels.
Read more in the User Guide.
- Parameters
- kernel{‘knn’, ‘rbf’} or callable, default=’rbf’
String identifier for kernel function to use or the kernel function itself. Only ‘rbf’ and ‘knn’ strings are valid inputs. The function passed should take two inputs, each of shape (n_samples, n_features), and return a (n_samples, n_samples) shaped weight matrix.
- gammafloat, default=20
Parameter for rbf kernel.
- n_neighborsint, default=7
Parameter for knn kernel which is a strictly positive integer.
- alphafloat, default=0.2
Clamping factor. A value in (0, 1) that specifies the relative amount that an instance should adopt the information from its neighbors as opposed to its initial label. alpha=0 means keeping the initial label information; alpha=1 means replacing all initial information.
- max_iterint, default=30
Maximum number of iterations allowed.
- tolfloat, default=1e-3
Convergence tolerance: threshold to consider the system at steady state.
- n_jobsint, default=None
The number of parallel jobs to run.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.
- Attributes
- X_ndarray of shape (n_samples, n_features)
Input array.
- classes_ndarray of shape (n_classes,)
The distinct labels used in classifying instances.
- label_distributions_ndarray of shape (n_samples, n_classes)
Categorical distribution for each item.
- transduction_ndarray of shape (n_samples,)
Label assigned to each item via the transduction.
- n_iter_int
Number of iterations run.
See also
LabelPropagationUnregularized graph based semi-supervised learning
References
Dengyong Zhou, Olivier Bousquet, Thomas Navin Lal, Jason Weston, Bernhard Schoelkopf. Learning with local and global consistency (2004) http://citeseer.ist.psu.edu/viewdoc/summary?doi=10.1.1.115.3219
Examples
>>> import numpy as np >>> from sklearn import datasets >>> from sklearn.semi_supervised import LabelSpreading >>> label_prop_model = LabelSpreading() >>> iris = datasets.load_iris() >>> rng = np.random.RandomState(42) >>> random_unlabeled_points = rng.rand(len(iris.target)) < 0.3 >>> labels = np.copy(iris.target) >>> labels[random_unlabeled_points] = -1 >>> label_prop_model.fit(iris.data, labels) LabelSpreading(...)
Methods
fit(X, y)Fit a semi-supervised label propagation model based
get_params([deep])Get parameters for this estimator.
predict(X)Performs inductive inference across the model.
Predict probability for each possible outcome.
score(X, y[, sample_weight])Return the mean accuracy on the given test data and labels.
set_params(**params)Set the parameters of this estimator.
-
__init__(kernel='rbf', *, gamma=20, n_neighbors=7, alpha=0.2, max_iter=30, tol=0.001, n_jobs=None)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(X, y)[source]¶ Fit a semi-supervised label propagation model based
All the input data is provided matrix X (labeled and unlabeled) and corresponding label matrix y with a dedicated marker value for unlabeled samples.
- Parameters
- Xarray-like of shape (n_samples, n_features)
A matrix of shape (n_samples, n_samples) will be created from this.
- yarray-like of shape (n_samples,)
n_labeled_samples(unlabeled points are marked as -1) All unlabeled samples will be transductively assigned labels.
- Returns
- selfobject
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsmapping of string to any
Parameter names mapped to their values.
-
predict(X)[source]¶ Performs inductive inference across the model.
- Parameters
- Xarray-like of shape (n_samples, n_features)
The data matrix.
- Returns
- yndarray of shape (n_samples,)
Predictions for input data.
-
predict_proba(X)[source]¶ Predict probability for each possible outcome.
Compute the probability estimates for each single sample in X and each possible outcome seen during training (categorical distribution).
- Parameters
- Xarray-like of shape (n_samples, n_features)
The data matrix.
- Returns
- probabilitiesndarray of shape (n_samples, n_classes)
Normalized probability distributions across class labels.
-
score(X, y, sample_weight=None)[source]¶ Return the mean accuracy on the given test data and labels.
In multi-label classification, this is the subset accuracy which is a harsh metric since you require for each sample that each label set be correctly predicted.
- Parameters
- Xarray-like of shape (n_samples, n_features)
Test samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True labels for X.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns
- scorefloat
Mean accuracy of self.predict(X) wrt. y.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfobject
Estimator instance.