sklearn.cluster.SpectralCoclustering¶
-
class
sklearn.cluster.SpectralCoclustering(n_clusters=3, *, svd_method='randomized', n_svd_vecs=None, mini_batch=False, init='k-means++', n_init=10, n_jobs='deprecated', random_state=None)[source]¶ Spectral Co-Clustering algorithm (Dhillon, 2001).
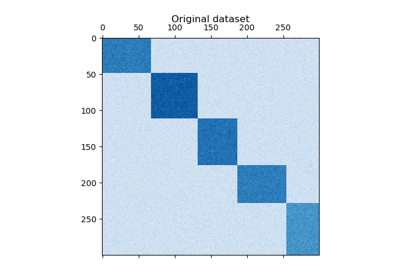
Clusters rows and columns of an array
Xto solve the relaxed normalized cut of the bipartite graph created fromXas follows: the edge between row vertexiand column vertexjhas weightX[i, j].The resulting bicluster structure is block-diagonal, since each row and each column belongs to exactly one bicluster.
Supports sparse matrices, as long as they are nonnegative.
Read more in the User Guide.
- Parameters
- n_clustersint, default=3
The number of biclusters to find.
- svd_method{‘randomized’, ‘arpack’}, default=’randomized’
Selects the algorithm for finding singular vectors. May be ‘randomized’ or ‘arpack’. If ‘randomized’, use
sklearn.utils.extmath.randomized_svd, which may be faster for large matrices. If ‘arpack’, usescipy.sparse.linalg.svds, which is more accurate, but possibly slower in some cases.- n_svd_vecsint, default=None
Number of vectors to use in calculating the SVD. Corresponds to
ncvwhensvd_method=arpackandn_oversampleswhensvd_methodis ‘randomized`.- mini_batchbool, default=False
Whether to use mini-batch k-means, which is faster but may get different results.
- init{‘k-means++’, ‘random’, or ndarray of shape (n_clusters, n_features), default=’k-means++’
Method for initialization of k-means algorithm; defaults to ‘k-means++’.
- n_initint, default=10
Number of random initializations that are tried with the k-means algorithm.
If mini-batch k-means is used, the best initialization is chosen and the algorithm runs once. Otherwise, the algorithm is run for each initialization and the best solution chosen.
- n_jobsint, default=None
The number of jobs to use for the computation. This works by breaking down the pairwise matrix into n_jobs even slices and computing them in parallel.
Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.Deprecated since version 0.23:
n_jobswas deprecated in version 0.23 and will be removed in 0.25.- random_stateint, RandomState instance, default=None
Used for randomizing the singular value decomposition and the k-means initialization. Use an int to make the randomness deterministic. See Glossary.
- Attributes
- rows_array-like of shape (n_row_clusters, n_rows)
Results of the clustering.
rows[i, r]is True if clustericontains rowr. Available only after callingfit.- columns_array-like of shape (n_column_clusters, n_columns)
Results of the clustering, like
rows.- row_labels_array-like of shape (n_rows,)
The bicluster label of each row.
- column_labels_array-like of shape (n_cols,)
The bicluster label of each column.
References
Dhillon, Inderjit S, 2001. Co-clustering documents and words using bipartite spectral graph partitioning.
Examples
>>> from sklearn.cluster import SpectralCoclustering >>> import numpy as np >>> X = np.array([[1, 1], [2, 1], [1, 0], ... [4, 7], [3, 5], [3, 6]]) >>> clustering = SpectralCoclustering(n_clusters=2, random_state=0).fit(X) >>> clustering.row_labels_ #doctest: +SKIP array([0, 1, 1, 0, 0, 0], dtype=int32) >>> clustering.column_labels_ #doctest: +SKIP array([0, 0], dtype=int32) >>> clustering SpectralCoclustering(n_clusters=2, random_state=0)
Methods
fit(X[, y])Creates a biclustering for X.
get_indices(i)Row and column indices of the i’th bicluster.
get_params([deep])Get parameters for this estimator.
get_shape(i)Shape of the i’th bicluster.
get_submatrix(i, data)Return the submatrix corresponding to bicluster
i.set_params(**params)Set the parameters of this estimator.
-
__init__(n_clusters=3, *, svd_method='randomized', n_svd_vecs=None, mini_batch=False, init='k-means++', n_init=10, n_jobs='deprecated', random_state=None)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
property
biclusters_¶ Convenient way to get row and column indicators together.
Returns the
rows_andcolumns_members.
-
fit(X, y=None)[source]¶ Creates a biclustering for X.
- Parameters
- Xarray-like, shape (n_samples, n_features)
- yIgnored
-
get_indices(i)[source]¶ Row and column indices of the i’th bicluster.
Only works if
rows_andcolumns_attributes exist.- Parameters
- iint
The index of the cluster.
- Returns
- row_indndarray, dtype=np.intp
Indices of rows in the dataset that belong to the bicluster.
- col_indndarray, dtype=np.intp
Indices of columns in the dataset that belong to the bicluster.
-
get_params(deep=True)[source]¶ Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsmapping of string to any
Parameter names mapped to their values.
-
get_shape(i)[source]¶ Shape of the i’th bicluster.
- Parameters
- iint
The index of the cluster.
- Returns
- shapetuple (int, int)
Number of rows and columns (resp.) in the bicluster.
-
get_submatrix(i, data)[source]¶ Return the submatrix corresponding to bicluster
i.- Parameters
- iint
The index of the cluster.
- dataarray-like
The data.
- Returns
- submatrixndarray
The submatrix corresponding to bicluster i.
Notes
Works with sparse matrices. Only works if
rows_andcolumns_attributes exist.
-
set_params(**params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfobject
Estimator instance.