Note
Click here to download the full example code or to run this example in your browser via Binder
Model Complexity Influence¶
Demonstrate how model complexity influences both prediction accuracy and computational performance.
The dataset is the Boston Housing dataset (resp. 20 Newsgroups) for regression (resp. classification).
For each class of models we make the model complexity vary through the choice of relevant model parameters and measure the influence on both computational performance (latency) and predictive power (MSE or Hamming Loss).
Out:
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.25, loss='modified_huber',
penalty='elasticnet')
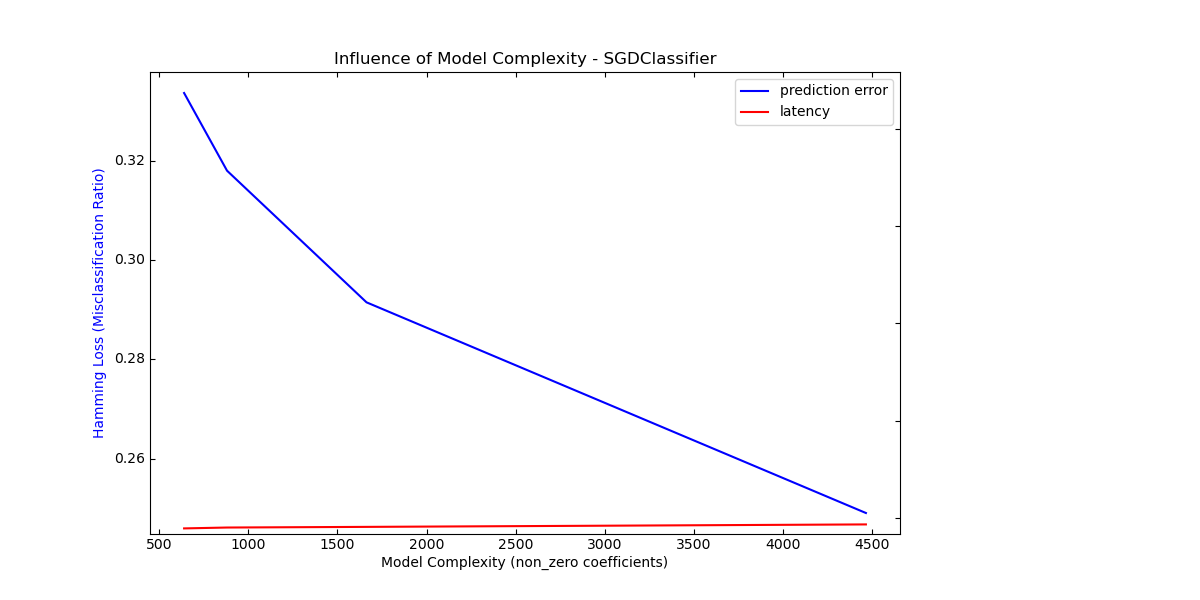
Complexity: 4466 | Hamming Loss (Misclassification Ratio): 0.2491 | Pred. Time: 0.020736s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.5, loss='modified_huber',
penalty='elasticnet')
Complexity: 1663 | Hamming Loss (Misclassification Ratio): 0.2915 | Pred. Time: 0.015413s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.75, loss='modified_huber',
penalty='elasticnet')
Complexity: 880 | Hamming Loss (Misclassification Ratio): 0.3180 | Pred. Time: 0.014005s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.9, loss='modified_huber',
penalty='elasticnet')
Complexity: 639 | Hamming Loss (Misclassification Ratio): 0.3337 | Pred. Time: 0.012106s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.1)
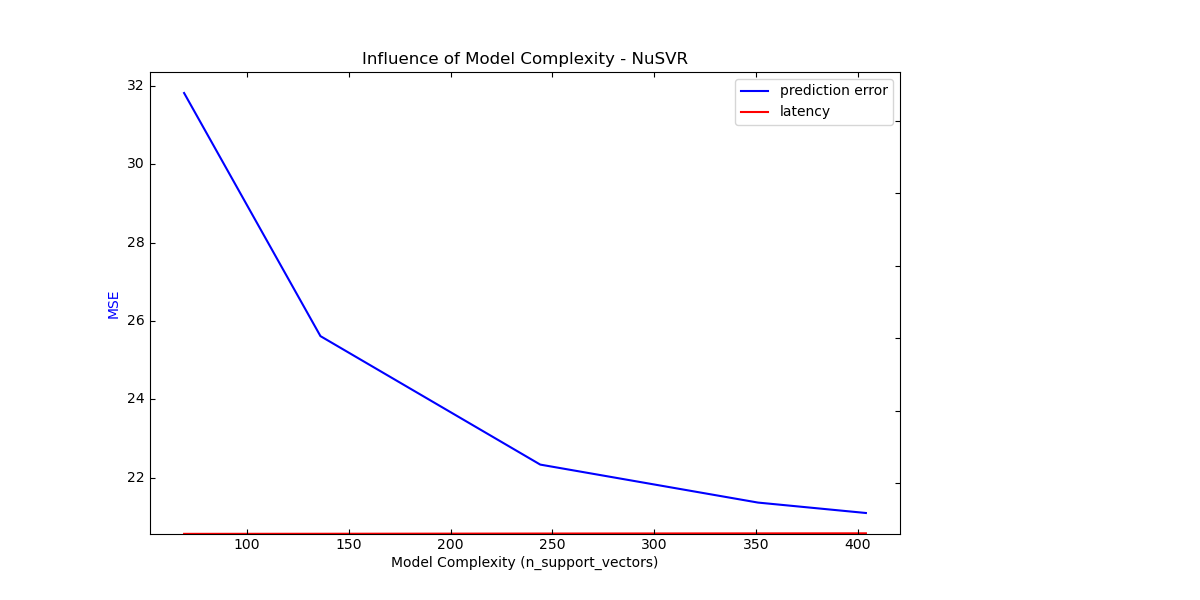
Complexity: 69 | MSE: 31.8139 | Pred. Time: 0.000318s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.25)
Complexity: 136 | MSE: 25.6140 | Pred. Time: 0.000514s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05)
Complexity: 244 | MSE: 22.3375 | Pred. Time: 0.000994s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.75)
Complexity: 351 | MSE: 21.3688 | Pred. Time: 0.001338s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.9)
Complexity: 404 | MSE: 21.1033 | Pred. Time: 0.001476s
Benchmarking GradientBoostingRegressor(n_estimators=10)
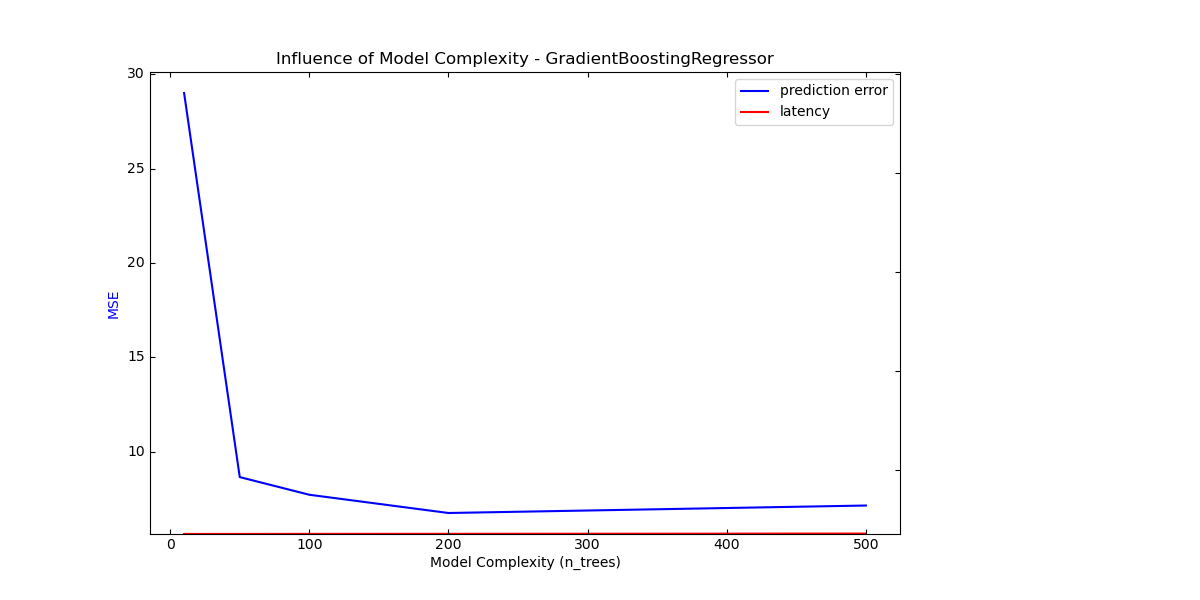
Complexity: 10 | MSE: 29.0148 | Pred. Time: 0.000113s
Benchmarking GradientBoostingRegressor(n_estimators=50)
Complexity: 50 | MSE: 8.6545 | Pred. Time: 0.000186s
Benchmarking GradientBoostingRegressor()
Complexity: 100 | MSE: 7.7179 | Pred. Time: 0.000263s
Benchmarking GradientBoostingRegressor(n_estimators=200)
Complexity: 200 | MSE: 6.7507 | Pred. Time: 0.000398s
Benchmarking GradientBoostingRegressor(n_estimators=500)
Complexity: 500 | MSE: 7.1471 | Pred. Time: 0.000961s
print(__doc__)
# Author: Eustache Diemert <eustache@diemert.fr>
# License: BSD 3 clause
import time
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1.parasite_axes import host_subplot
from mpl_toolkits.axisartist.axislines import Axes
from scipy.sparse.csr import csr_matrix
from sklearn import datasets
from sklearn.utils import shuffle
from sklearn.metrics import mean_squared_error
from sklearn.svm import NuSVR
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import hamming_loss
# #############################################################################
# Routines
# Initialize random generator
np.random.seed(0)
def generate_data(case, sparse=False):
"""Generate regression/classification data."""
if case == 'regression':
X, y = datasets.load_boston(return_X_y=True)
elif case == 'classification':
X, y = datasets.fetch_20newsgroups_vectorized(subset='all',
return_X_y=True)
X, y = shuffle(X, y)
offset = int(X.shape[0] * 0.8)
X_train, y_train = X[:offset], y[:offset]
X_test, y_test = X[offset:], y[offset:]
if sparse:
X_train = csr_matrix(X_train)
X_test = csr_matrix(X_test)
else:
X_train = np.array(X_train)
X_test = np.array(X_test)
y_test = np.array(y_test)
y_train = np.array(y_train)
data = {'X_train': X_train, 'X_test': X_test, 'y_train': y_train,
'y_test': y_test}
return data
def benchmark_influence(conf):
"""
Benchmark influence of :changing_param: on both MSE and latency.
"""
prediction_times = []
prediction_powers = []
complexities = []
for param_value in conf['changing_param_values']:
conf['tuned_params'][conf['changing_param']] = param_value
estimator = conf['estimator'](**conf['tuned_params'])
print("Benchmarking %s" % estimator)
estimator.fit(conf['data']['X_train'], conf['data']['y_train'])
conf['postfit_hook'](estimator)
complexity = conf['complexity_computer'](estimator)
complexities.append(complexity)
start_time = time.time()
for _ in range(conf['n_samples']):
y_pred = estimator.predict(conf['data']['X_test'])
elapsed_time = (time.time() - start_time) / float(conf['n_samples'])
prediction_times.append(elapsed_time)
pred_score = conf['prediction_performance_computer'](
conf['data']['y_test'], y_pred)
prediction_powers.append(pred_score)
print("Complexity: %d | %s: %.4f | Pred. Time: %fs\n" % (
complexity, conf['prediction_performance_label'], pred_score,
elapsed_time))
return prediction_powers, prediction_times, complexities
def plot_influence(conf, mse_values, prediction_times, complexities):
"""
Plot influence of model complexity on both accuracy and latency.
"""
plt.figure(figsize=(12, 6))
host = host_subplot(111, axes_class=Axes)
plt.subplots_adjust(right=0.75)
par1 = host.twinx()
host.set_xlabel('Model Complexity (%s)' % conf['complexity_label'])
y1_label = conf['prediction_performance_label']
y2_label = "Time (s)"
host.set_ylabel(y1_label)
par1.set_ylabel(y2_label)
p1, = host.plot(complexities, mse_values, 'b-', label="prediction error")
p2, = par1.plot(complexities, prediction_times, 'r-',
label="latency")
host.legend(loc='upper right')
host.axis["left"].label.set_color(p1.get_color())
par1.axis["right"].label.set_color(p2.get_color())
plt.title('Influence of Model Complexity - %s' % conf['estimator'].__name__)
plt.show()
def _count_nonzero_coefficients(estimator):
a = estimator.coef_.toarray()
return np.count_nonzero(a)
# #############################################################################
# Main code
regression_data = generate_data('regression')
classification_data = generate_data('classification', sparse=True)
configurations = [
{'estimator': SGDClassifier,
'tuned_params': {'penalty': 'elasticnet', 'alpha': 0.001, 'loss':
'modified_huber', 'fit_intercept': True, 'tol': 1e-3},
'changing_param': 'l1_ratio',
'changing_param_values': [0.25, 0.5, 0.75, 0.9],
'complexity_label': 'non_zero coefficients',
'complexity_computer': _count_nonzero_coefficients,
'prediction_performance_computer': hamming_loss,
'prediction_performance_label': 'Hamming Loss (Misclassification Ratio)',
'postfit_hook': lambda x: x.sparsify(),
'data': classification_data,
'n_samples': 30},
{'estimator': NuSVR,
'tuned_params': {'C': 1e3, 'gamma': 2 ** -15},
'changing_param': 'nu',
'changing_param_values': [0.1, 0.25, 0.5, 0.75, 0.9],
'complexity_label': 'n_support_vectors',
'complexity_computer': lambda x: len(x.support_vectors_),
'data': regression_data,
'postfit_hook': lambda x: x,
'prediction_performance_computer': mean_squared_error,
'prediction_performance_label': 'MSE',
'n_samples': 30},
{'estimator': GradientBoostingRegressor,
'tuned_params': {'loss': 'ls'},
'changing_param': 'n_estimators',
'changing_param_values': [10, 50, 100, 200, 500],
'complexity_label': 'n_trees',
'complexity_computer': lambda x: x.n_estimators,
'data': regression_data,
'postfit_hook': lambda x: x,
'prediction_performance_computer': mean_squared_error,
'prediction_performance_label': 'MSE',
'n_samples': 30},
]
for conf in configurations:
prediction_performances, prediction_times, complexities = \
benchmark_influence(conf)
plot_influence(conf, prediction_performances, prediction_times,
complexities)
Total running time of the script: ( 0 minutes 23.358 seconds)