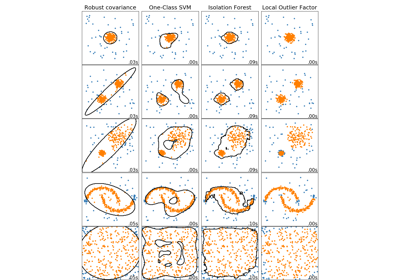
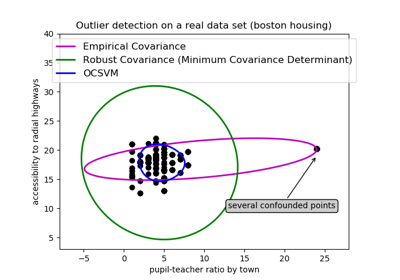
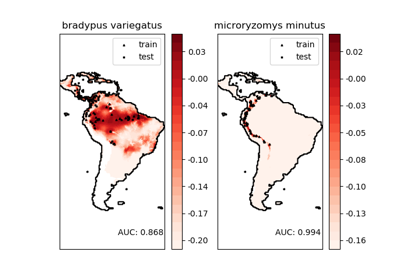
sklearn.svm.OneClassSVM¶
-
class
sklearn.svm.OneClassSVM(kernel=’rbf’, degree=3, gamma=’auto_deprecated’, coef0=0.0, tol=0.001, nu=0.5, shrinking=True, cache_size=200, verbose=False, max_iter=-1, random_state=None)[source]¶ Unsupervised Outlier Detection.
Estimate the support of a high-dimensional distribution.
The implementation is based on libsvm.
Read more in the User Guide.
Parameters: - kernel : string, optional (default=’rbf’)
Specifies the kernel type to be used in the algorithm. It must be one of ‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’, ‘precomputed’ or a callable. If none is given, ‘rbf’ will be used. If a callable is given it is used to precompute the kernel matrix.
- degree : int, optional (default=3)
Degree of the polynomial kernel function (‘poly’). Ignored by all other kernels.
- gamma : float, optional (default=’auto’)
Kernel coefficient for ‘rbf’, ‘poly’ and ‘sigmoid’.
Current default is ‘auto’ which uses 1 / n_features, if
gamma='scale'is passed then it uses 1 / (n_features * X.var()) as value of gamma. The current default of gamma, ‘auto’, will change to ‘scale’ in version 0.22. ‘auto_deprecated’, a deprecated version of ‘auto’ is used as a default indicating that no explicit value of gamma was passed.- coef0 : float, optional (default=0.0)
Independent term in kernel function. It is only significant in ‘poly’ and ‘sigmoid’.
- tol : float, optional
Tolerance for stopping criterion.
- nu : float, optional
An upper bound on the fraction of training errors and a lower bound of the fraction of support vectors. Should be in the interval (0, 1]. By default 0.5 will be taken.
- shrinking : boolean, optional
Whether to use the shrinking heuristic.
- cache_size : float, optional
Specify the size of the kernel cache (in MB).
- verbose : bool, default: False
Enable verbose output. Note that this setting takes advantage of a per-process runtime setting in libsvm that, if enabled, may not work properly in a multithreaded context.
- max_iter : int, optional (default=-1)
Hard limit on iterations within solver, or -1 for no limit.
- random_state : int, RandomState instance or None, optional (default=None)
Ignored.
Deprecated since version 0.20:
random_statehas been deprecated in 0.20 and will be removed in 0.22.
Attributes: - support_ : array-like, shape = [n_SV]
Indices of support vectors.
- support_vectors_ : array-like, shape = [nSV, n_features]
Support vectors.
- dual_coef_ : array, shape = [1, n_SV]
Coefficients of the support vectors in the decision function.
- coef_ : array, shape = [1, n_features]
Weights assigned to the features (coefficients in the primal problem). This is only available in the case of a linear kernel.
coef_ is readonly property derived from
dual_coef_andsupport_vectors_- intercept_ : array, shape = [1,]
Constant in the decision function.
- offset_ : float
Offset used to define the decision function from the raw scores. We have the relation: decision_function = score_samples -
offset_. The offset is the opposite ofintercept_and is provided for consistency with other outlier detection algorithms.
Examples
>>> from sklearn.svm import OneClassSVM >>> X = [[0], [0.44], [0.45], [0.46], [1]] >>> clf = OneClassSVM(gamma='auto').fit(X) >>> clf.predict(X) array([-1, 1, 1, 1, -1]) >>> clf.score_samples(X) array([1.7798..., 2.0547..., 2.0556..., 2.0561..., 1.7332...])
Methods
decision_function(self, X)Signed distance to the separating hyperplane. fit(self, X[, y, sample_weight])Detects the soft boundary of the set of samples X. fit_predict(self, X[, y])Performs fit on X and returns labels for X. get_params(self[, deep])Get parameters for this estimator. predict(self, X)Perform classification on samples in X. score_samples(self, X)Raw scoring function of the samples. set_params(self, \*\*params)Set the parameters of this estimator. -
__init__(self, kernel=’rbf’, degree=3, gamma=’auto_deprecated’, coef0=0.0, tol=0.001, nu=0.5, shrinking=True, cache_size=200, verbose=False, max_iter=-1, random_state=None)[source]¶
-
decision_function(self, X)[source]¶ Signed distance to the separating hyperplane.
Signed distance is positive for an inlier and negative for an outlier.
Parameters: - X : array-like, shape (n_samples, n_features)
Returns: - dec : array-like, shape (n_samples,)
Returns the decision function of the samples.
-
fit(self, X, y=None, sample_weight=None, **params)[source]¶ Detects the soft boundary of the set of samples X.
Parameters: - X : {array-like, sparse matrix}, shape (n_samples, n_features)
Set of samples, where n_samples is the number of samples and n_features is the number of features.
- sample_weight : array-like, shape (n_samples,)
Per-sample weights. Rescale C per sample. Higher weights force the classifier to put more emphasis on these points.
- y : Ignored
not used, present for API consistency by convention.
Returns: - self : object
Notes
If X is not a C-ordered contiguous array it is copied.
-
fit_predict(self, X, y=None)[source]¶ Performs fit on X and returns labels for X.
Returns -1 for outliers and 1 for inliers.
Parameters: - X : ndarray, shape (n_samples, n_features)
Input data.
- y : Ignored
not used, present for API consistency by convention.
Returns: - y : ndarray, shape (n_samples,)
1 for inliers, -1 for outliers.
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
predict(self, X)[source]¶ Perform classification on samples in X.
For a one-class model, +1 or -1 is returned.
Parameters: - X : {array-like, sparse matrix}, shape (n_samples, n_features)
For kernel=”precomputed”, the expected shape of X is [n_samples_test, n_samples_train]
Returns: - y_pred : array, shape (n_samples,)
Class labels for samples in X.
-
score_samples(self, X)[source]¶ Raw scoring function of the samples.
Parameters: - X : array-like, shape (n_samples, n_features)
Returns: - score_samples : array-like, shape (n_samples,)
Returns the (unshifted) scoring function of the samples.
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self