sklearn.preprocessing.PowerTransformer¶
-
class
sklearn.preprocessing.PowerTransformer(method=’yeo-johnson’, standardize=True, copy=True)[source]¶ Apply a power transform featurewise to make data more Gaussian-like.
Power transforms are a family of parametric, monotonic transformations that are applied to make data more Gaussian-like. This is useful for modeling issues related to heteroscedasticity (non-constant variance), or other situations where normality is desired.
Currently, PowerTransformer supports the Box-Cox transform and the Yeo-Johnson transform. The optimal parameter for stabilizing variance and minimizing skewness is estimated through maximum likelihood.
Box-Cox requires input data to be strictly positive, while Yeo-Johnson supports both positive or negative data.
By default, zero-mean, unit-variance normalization is applied to the transformed data.
Read more in the User Guide.
Parameters: - method : str, (default=’yeo-johnson’)
The power transform method. Available methods are:
- ‘yeo-johnson’ [Rf3e1504535de-1], works with positive and negative values
- ‘box-cox’ [Rf3e1504535de-2], only works with strictly positive values
- standardize : boolean, default=True
Set to True to apply zero-mean, unit-variance normalization to the transformed output.
- copy : boolean, optional, default=True
Set to False to perform inplace computation during transformation.
Attributes: - lambdas_ : array of float, shape (n_features,)
The parameters of the power transformation for the selected features.
See also
power_transform- Equivalent function without the estimator API.
QuantileTransformer- Maps data to a standard normal distribution with the parameter
output_distribution='normal'.
Notes
NaNs are treated as missing values: disregarded in
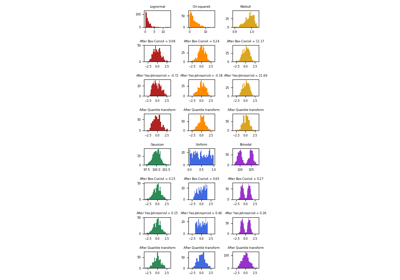
fit, and maintained intransform.For a comparison of the different scalers, transformers, and normalizers, see examples/preprocessing/plot_all_scaling.py.
References
[Rf3e1504535de-1] I.K. Yeo and R.A. Johnson, “A new family of power transformations to improve normality or symmetry.” Biometrika, 87(4), pp.954-959, (2000). [Rf3e1504535de-2] G.E.P. Box and D.R. Cox, “An Analysis of Transformations”, Journal of the Royal Statistical Society B, 26, 211-252 (1964). Examples
>>> import numpy as np >>> from sklearn.preprocessing import PowerTransformer >>> pt = PowerTransformer() >>> data = [[1, 2], [3, 2], [4, 5]] >>> print(pt.fit(data)) PowerTransformer(copy=True, method='yeo-johnson', standardize=True) >>> print(pt.lambdas_) [ 1.386... -3.100...] >>> print(pt.transform(data)) [[-1.316... -0.707...] [ 0.209... -0.707...] [ 1.106... 1.414...]]
Methods
fit(self, X[, y])Estimate the optimal parameter lambda for each feature. fit_transform(self, X[, y])get_params(self[, deep])Get parameters for this estimator. inverse_transform(self, X)Apply the inverse power transformation using the fitted lambdas. set_params(self, \*\*params)Set the parameters of this estimator. transform(self, X)Apply the power transform to each feature using the fitted lambdas. -
fit(self, X, y=None)[source]¶ Estimate the optimal parameter lambda for each feature.
The optimal lambda parameter for minimizing skewness is estimated on each feature independently using maximum likelihood.
Parameters: - X : array-like, shape (n_samples, n_features)
The data used to estimate the optimal transformation parameters.
- y : Ignored
Returns: - self : object
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
inverse_transform(self, X)[source]¶ Apply the inverse power transformation using the fitted lambdas.
The inverse of the Box-Cox transformation is given by:
if lambda == 0: X = exp(X_trans) else: X = (X_trans * lambda + 1) ** (1 / lambda)
The inverse of the Yeo-Johnson transformation is given by:
if X >= 0 and lambda == 0: X = exp(X_trans) - 1 elif X >= 0 and lambda != 0: X = (X_trans * lambda + 1) ** (1 / lambda) - 1 elif X < 0 and lambda != 2: X = 1 - (-(2 - lambda) * X_trans + 1) ** (1 / (2 - lambda)) elif X < 0 and lambda == 2: X = 1 - exp(-X_trans)
Parameters: - X : array-like, shape (n_samples, n_features)
The transformed data.
Returns: - X : array-like, shape (n_samples, n_features)
The original data
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self