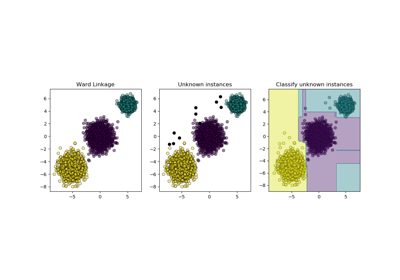
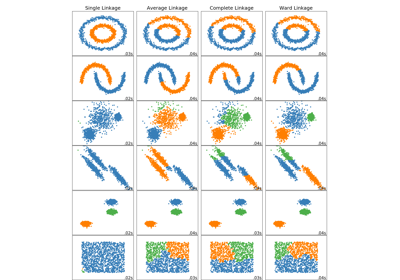
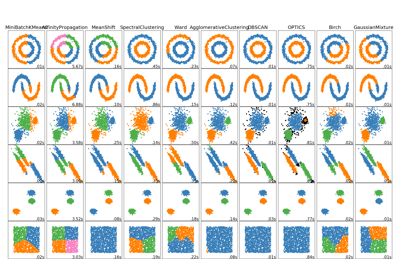
sklearn.cluster.AgglomerativeClustering¶
-
class
sklearn.cluster.AgglomerativeClustering(n_clusters=2, affinity=’euclidean’, memory=None, connectivity=None, compute_full_tree=’auto’, linkage=’ward’, pooling_func=’deprecated’, distance_threshold=None)[source]¶ Agglomerative Clustering
Recursively merges the pair of clusters that minimally increases a given linkage distance.
Read more in the User Guide.
Parameters: - n_clusters : int or None, optional (default=2)
The number of clusters to find. It must be
Noneifdistance_thresholdis notNone.- affinity : string or callable, default: “euclidean”
Metric used to compute the linkage. Can be “euclidean”, “l1”, “l2”, “manhattan”, “cosine”, or “precomputed”. If linkage is “ward”, only “euclidean” is accepted. If “precomputed”, a distance matrix (instead of a similarity matrix) is needed as input for the fit method.
- memory : None, str or object with the joblib.Memory interface, optional
Used to cache the output of the computation of the tree. By default, no caching is done. If a string is given, it is the path to the caching directory.
- connectivity : array-like or callable, optional
Connectivity matrix. Defines for each sample the neighboring samples following a given structure of the data. This can be a connectivity matrix itself or a callable that transforms the data into a connectivity matrix, such as derived from kneighbors_graph. Default is None, i.e, the hierarchical clustering algorithm is unstructured.
- compute_full_tree : bool or ‘auto’ (optional)
Stop early the construction of the tree at n_clusters. This is useful to decrease computation time if the number of clusters is not small compared to the number of samples. This option is useful only when specifying a connectivity matrix. Note also that when varying the number of clusters and using caching, it may be advantageous to compute the full tree. It must be
Trueifdistance_thresholdis notNone.- linkage : {“ward”, “complete”, “average”, “single”}, optional (default=”ward”)
Which linkage criterion to use. The linkage criterion determines which distance to use between sets of observation. The algorithm will merge the pairs of cluster that minimize this criterion.
- ward minimizes the variance of the clusters being merged.
- average uses the average of the distances of each observation of the two sets.
- complete or maximum linkage uses the maximum distances between all observations of the two sets.
- single uses the minimum of the distances between all observations of the two sets.
- pooling_func : callable, default=’deprecated’
Ignored.
Deprecated since version 0.20:
pooling_funchas been deprecated in 0.20 and will be removed in 0.22.- distance_threshold : float, optional (default=None)
The linkage distance threshold above which, clusters will not be merged. If not
None,n_clustersmust beNoneandcompute_full_treemust beTrue.New in version 0.21.
Attributes: - n_clusters_ : int
The number of clusters found by the algorithm. If
distance_threshold=None, it will be equal to the givenn_clusters.- labels_ : array [n_samples]
cluster labels for each point
- n_leaves_ : int
Number of leaves in the hierarchical tree.
- n_connected_components_ : int
The estimated number of connected components in the graph.
- children_ : array-like, shape (n_samples-1, 2)
The children of each non-leaf node. Values less than n_samples correspond to leaves of the tree which are the original samples. A node
igreater than or equal to n_samples is a non-leaf node and has childrenchildren_[i - n_samples]. Alternatively at the i-th iteration, children[i][0] and children[i][1] are merged to form noden_samples + i
Examples
>>> from sklearn.cluster import AgglomerativeClustering >>> import numpy as np >>> X = np.array([[1, 2], [1, 4], [1, 0], ... [4, 2], [4, 4], [4, 0]]) >>> clustering = AgglomerativeClustering().fit(X) >>> clustering AgglomerativeClustering(affinity='euclidean', compute_full_tree='auto', connectivity=None, distance_threshold=None, linkage='ward', memory=None, n_clusters=2, pooling_func='deprecated') >>> clustering.labels_ array([1, 1, 1, 0, 0, 0])
Methods
fit(self, X[, y])Fit the hierarchical clustering on the data fit_predict(self, X[, y])Performs clustering on X and returns cluster labels. get_params(self[, deep])Get parameters for this estimator. set_params(self, \*\*params)Set the parameters of this estimator. -
__init__(self, n_clusters=2, affinity=’euclidean’, memory=None, connectivity=None, compute_full_tree=’auto’, linkage=’ward’, pooling_func=’deprecated’, distance_threshold=None)[source]¶
-
fit(self, X, y=None)[source]¶ Fit the hierarchical clustering on the data
Parameters: - X : array-like, shape = [n_samples, n_features]
Training data. Shape [n_samples, n_features], or [n_samples, n_samples] if affinity==’precomputed’.
- y : Ignored
Returns: - self
-
fit_predict(self, X, y=None)[source]¶ Performs clustering on X and returns cluster labels.
Parameters: - X : ndarray, shape (n_samples, n_features)
Input data.
- y : Ignored
not used, present for API consistency by convention.
Returns: - labels : ndarray, shape (n_samples,)
cluster labels
-
get_params(self, deep=True)[source]¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
set_params(self, **params)[source]¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self